Comparative Study of Envelope Proteins of Dengue Virus of All Four Serotypes Isolated In India
Sumanta Dey1, Papiya Nandy1, Sukhen Das1,2, Ashesh Nandy1*
Affiliation
- 1Centre for Interdisciplinary Research and Education, Kolkata, India
- 2Physics Dept., Jadavpur University, Jadavpur, Kolkata, India
Corresponding Author
Ashesh Nandy. Centre for Interdisciplinary Research and Education, 404B Jodhpur Park, Kolkata 700068, India, E-mail: anandy43@yahoo.com
Citation
Nandy, A., et al. Comparative Study of Envelope Proteins of Dengue Virus of All Four Serotypes Isolated In India. (2016) Bioinfo Proteom Img Anal 2(2): 105- 113.
Copy rights
© 2016 Nandy, A. This is an Open access article distributed under the terms of Creative Commons Attribution 4.0 International License.
Keywords
Dengue serotypes; Dengue envelope phylogeny; 2D Graphical representation; Dengue transition/transversion ratios; Amino acid usage; Dengue epidemics in India; Peptide vaccines
Abstract
Dengue virus infections spread mainly by the Aedes mosquitoes, affect 50 to 100 million people annually, of which a small percentage can suffer severe dengue hemorrhagic fever that can be fatal, and dengue fever in epidemic form has been affecting mainly the tropical countries. The tragedy is compounded by the fact that there are as yet no drugs or vaccines to treat the disease or act as preventives. Also, dengue virus belongs to the Flavivirus family, which also includes the latest Zika virus that is also spread by the same vector. The impact of such viruses makes it important to understand and characterize them, especially their surface proteins which continue to remain the first point of attack for drug and prophylaxis design. In this paper we consider the dengue surface situated envelope protein for all dengue serotypes from India to characterize the sequences and determine their similarities and dissimilarities with a view to assist in monitoring viral changes and in design of drugs and vaccines. We found the nucleotide sequences of the envelope genes of all four dengue serotypes to be very similar at the 3´-end, while there are slightly more differences near the 5´-end where the presence of the glycosilation site is known to be accompanied by large variability in the amino acid sequences. Phylogenetic analysis showed that the dengue type 4 (DENV4) is cladistically slightly removed compared to the other three dengue serotypes (DENV1, DENV3 and DENV2). The codon usage profiles show differences in codon usage biases between various serotypes. All four serotypes exhibit similar transition-transversion ratios and amino acid usage patterns; hydropathy indexes show small differences between DENV1, 4 and DENV2, 3. Our analyses identified three surface exposed regions in the protein sequences that are common to all four serotypes, which may be of interest in vaccine design.
Introduction
??Dengue virus is vector borne, spread primarily by the bites of the Aedes group of mosquitoes. Clinical manifestations range from asymptomatic infection to a self-limiting febrile illness to, in severe cases, dengue hemorrhagic fever (DHF) that can lead to fatalities. In recent years the virus has spread widely following the growth of human population with about 2.5 billion people in risk-prone areas, mainly in tropical lands, and inadequate mosquito control. New incidences of dengue infections are estimated at around 50 - 100 million annually worldwide, due in part to spread of the virus through greater international travel and larger range for insects due to global warming, of which about half a million suffer from life-threatening complications[1]. There have been several dengue epidemics around the world leading to significant impact on human health and economy[2-4], but dengue research has not received the kind of attention and funds that would do justice to the gravity of the situation[2]. As of today there are no drugs and no licensed vaccines to treat the disease, neither are there diagnostic tools[5] although several are in various stages of development[6].
??The dengue virus belongs to the Flavivirus family, genus Flaviviridae, which includes other viruses like the Japanese encephalitis virus, the West Nile virus, the yellow fever virus, and the Zika virus. The genome is a positive sense single stranded RNA virus about 11000 base pairs long that codes for three structural proteins – the capsid C, membrane protein M and envelope protein E - and seven nonstructural proteins (NS1, NS2a, NS2b, NS3, NS4a, NS4b, NS5), and also includes short non-coding regions on both the 5? and 3? ends. Antigenic determinations recognize four serotypes labeled as types 1 to 4, designated as DENV1, DENV2, DENV3 and DENV4; a fifth serotype was recently reported[7] but much is not known about it yet. Another recent report[8] based on extensive research with many dengue viral strains found that rather than distinct clusters of differences between the antigenic structures of the several serotypes, these form rather a continuum, which makes vaccine design against the dengue more complicated. It is also of interest to note that dengue viral pathogenesis appears to depend upon the viral type[9], thus adding another layer of complexity to monitoring of changes in the viral genomes.
??Dengue in India is suspected to have first caused infections in 1780, but the first proven case of dengue fever (DF) epidemic was in 1963 - 64[6]. Since then DF epidemics have been reported in Delhi in 1967, in Kanpur in 1968, in Gujarat in 1988 - 89, in Delhi in 1996 and in several succeeding years. DHF was reported in India for the first time around 1988, with the first DHF epidemic around Delhi and Lucknow in 1996 and again in 2004 in northern India. In these epidemics, all four serotypes of the dengue virus, viz., DENV1, DENV2, DENV3 and DENV4, and their various genotypes have been found co-circulating, in different combinations. DENV4 was determined in Kanpur in 1966, and in the following year both DENV2 and DENV4 were isolated. DENV3 was isolated in South India around the same time and in the 1968 epidemic all four serotypes were reportedly determined. DENV2 was predominant in Delhi until 2003 when all four serotypes were found there, but since 2007 - 09 DENV1 seems to have predominated over DENV2 and DENV3[6]. The presence of several serotypes co-circulating in a locality present opportunity for concurrent infections in some patients leading to additional clinical complications.
??Dengue being a poorly researched disease yet with wide ranging effects constituting a global health threat thus requires understanding of its characteristics for any semblance of rational control, especially in a country as badly affected by the disease and with as large and dense a population as India. For vaccine design or drug development, the usual first choice is the surface proteins of the virus particle. From that point of view it is vital to understand the characteristics of the envelope protein of the dengue virion which exists as a homodimerlying flat on the virus totaling about 180 E-proteins on each virion[10]. While several reports have been published on clinical manifestations and experimental side of individual dengue types from various regions of India[6], a comprehensive characterization of the dengue envelope gene which could be crucial for any eventual drug or vaccine design does not seem to have been published so far.
??It might be appropriate to note here that due to lack of any definitive diagnostic tools for dengue, the disease can on occasion be unrecognized or be mistaken for chikingunya, another viral infection. The recent epidemic of Zika virus in Brazil and Latin American countries have raised further concerns since both, the dengue and the Zika viruses, belong to the Flavivirus family and are genetically similar. Both are spread by the same vector, and for the Zika virus too there are no drugs or vaccines yet; it is possible that rapid spread of the Zika virus can take place worldwide too and impact on countries hitherto remaining untouched. It is therefore important to understand as fully as possible a member of the Flavivirus family such as the dengue so we can better prepare for evolving eventualities. It is with these ideas in view that we have undertaken the following exercise.
Materials and Methods
??We have downloaded all 80 sequences of the envelope gene of all four types of the dengue virus that have circulated in India and are available in the GenBank database of National Institute of Health, USA, last accessed 15th May 2016; a summary is given in Table 1. They are all partial cds, but actually missing a few bases at 5?- and/or 3?-end. While normally we use complete cds’s for all our bioinformatics analyses, in the interest of doing as inclusive an analysis as possible for the Indian dengue viral sequences where complete cds’s are not available, we have used these partial sequences for all the analyses here with the proviso that some results might change, however slightly, once complete cds’s become available.
Table 1: Number of sequences of various dengue serotypes available in GenBank (accessed May 15, 2016) related to circulation in India.
| Dengue Type | No of Sequences |
| 1 | 23 |
| 2 | 19 |
| 3 | 17 |
| 4 | 21 |
??For comparison of gene sequences of the different dengue serotypes, we use a 2D graphical representation method that presents a visual rendering of the distribution of bases in a sequence. For the graphical representation, the sequences are plotted as a walk in a 2D rectangular grid taking one step in the negative x-direction for an adenine, in the positive y-direction for a cytosine, in the positive x-direction for a thymine/uracil and in the negative y-direction for a guanine. This yields (x, y) value for each base in a sequence and plots of the points of successive nucleotides generate a curve in the 2D space representing the distribution of bases along the sequence[11].
??Numerical characterization of the graphs generated in the 2D representation permit quantitative estimation of sequence similarities and distances between them[12]. Taking the weighted centre of mass of the graph of the selected sequence as ?x = ?xi/ N and ?y = ?yi/N where N is the total number of nucleotides of the segment, we can compute the distance (gR) from the origin to the centre of mass, referred to as the graph radius, as an index of the plot; differences between two sequences can be related to the distance between them computed through the differences between the graph radii. The gR has been shown[13] to be a sensitive measure of the distribution of bases in a segment; while the absolute values of the gR is irrelevant for our purposes, equalities of two gR values of two segments or gene sequences imply complete identity between the base distribution and composition in the two sequences. This property can be used to rapidly scan large numbers of sequences or sequence segments to find base distributions that are identical between them.
??Phylogenetic trees showing the evolutionary relationship between the sequences were drawn using the software MEGA5.2. We also calculated the transition/transversion ratio along with the amino acid composition of the envelope gene sequences by using the same software.
Results and Discussions
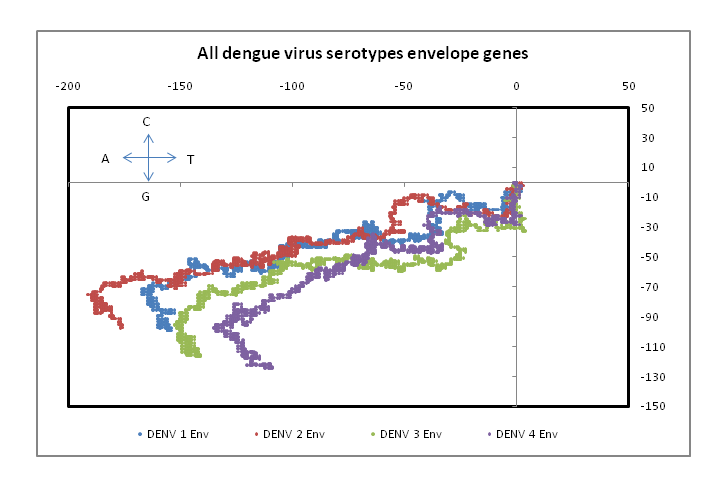
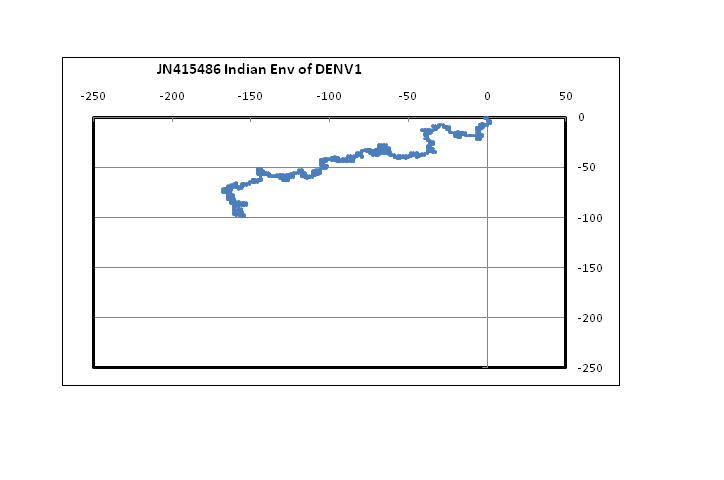
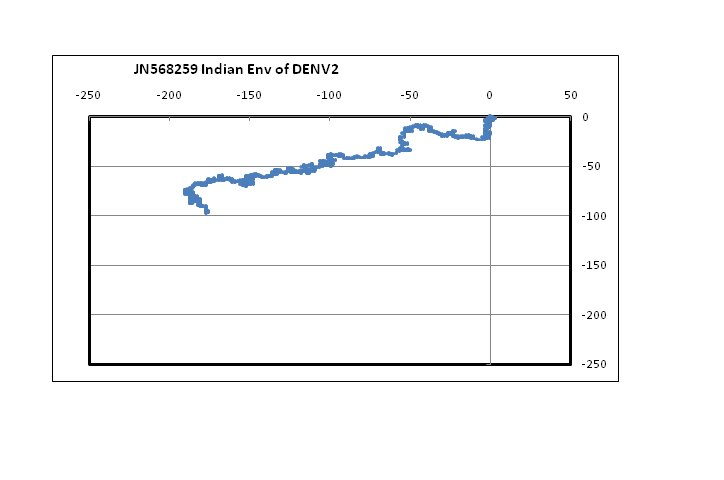
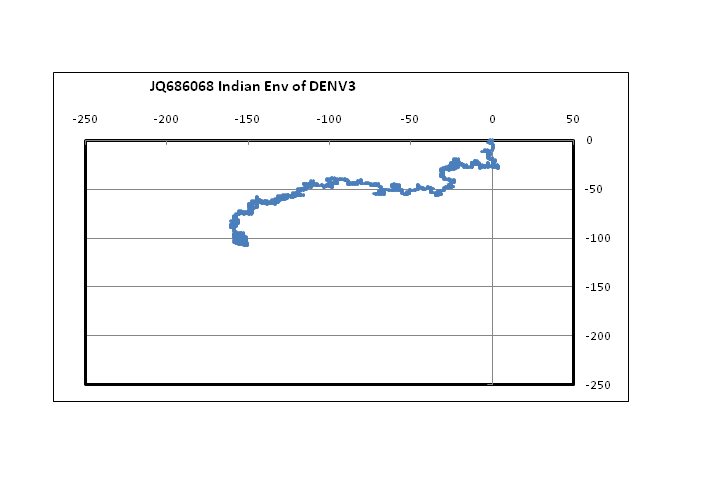
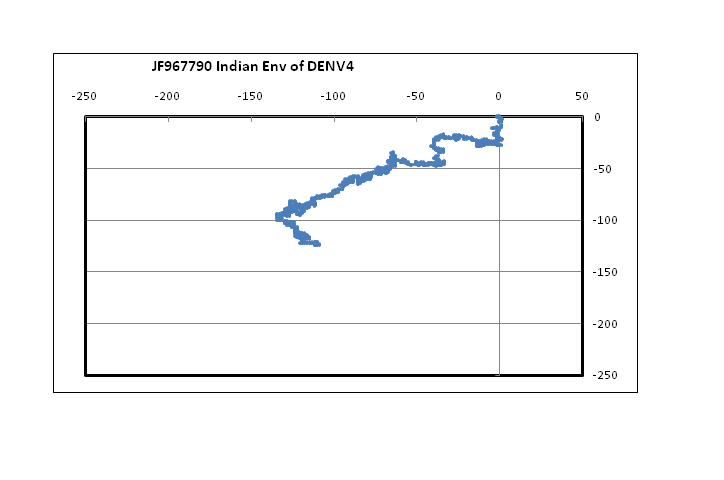
??Figure 1 shows plots in a 2D graphical representation scheme of one sample each of the envelope gene sequence of the four dengue serotypes. The sequences are plotted with the 5?-end at the origin; the plot ends with the 3?-end, in this case at around the centre of the graph. It is easy to see that serotypes 1, 2 and 3 are quite similar to each other; however, the type 4 dengue sequence plotted here appear to be comparatively more different. In the case of DENV2 we notice that the plot stretches more in the negative x-direction than the other dengue serotypes. This arises from comparatively higher percentage of adenines in DENV2, especially in the 5?-end, around the 121 - 200 nt region. In the case of protein sequences of these genes, there is large variation in the amino acids in the corresponding region, which is to be expected since this is adjacent to the glycosilation site[14]. It is to be noted, however, that for all four serotypes, the 3?-end constituting the stem and anchor region of the envelope protein are very similar showing that the sequence there should be quite important for the virus’s evolutionary changes. We had, in fact, observed a similar situation with influenza a hemagglutinin protein[15] where the second segment, HA2, stitches the hemagglutinin protein into the viral capsid and therefore appears to mutate less than the other segment, HA1. In the case of influenza A neuraminidase also, we had seen that the 3?-end of the neuraminidase protein was extremely stable, apparently due to the binding it makes with the neighboring protein in the quaternary structure[16]. Thus the 3?-end plays a vital role in viral surface protein stability, and this is evident in the case of the dengue envelope proteins also.
Figure 1: 2D graphical representations of envelope gene sequences of all four serotypes of the dengue virus circulating in India. The axes are as shown in the combined figure where all four envelope genes are plotted together (Blue – DENV1; Brown –DENV2; Green – DENV3; Violet – DENV4).
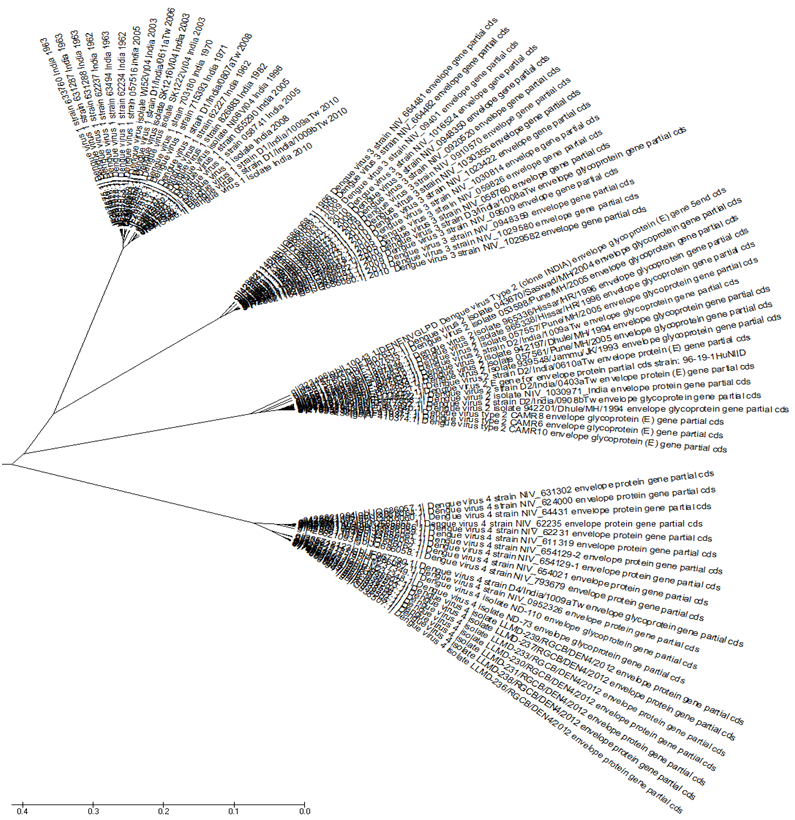
??In Figure 2 we address the problem of evolutionary relationships between the protein viral sequences collected in our database of Table 1. Using MEGA5.2 we first align the sequences and then work out the phylogenetic relationships. The phylogenetic tree in Figure 2 shows that the four serotypes fall into separate clades, each type belonging to its own clade. DENV1 and 3 clades are close by at the top of the figure; along with DENV2 they form one super clade. DENV4 belongs to a clade by itself, with two sub-clades, one for the period before 2000 AD and one after. In terms of ancestry, dengue type 4 would seem to be the most ancient of them all. It is interesting to note from the review by[6] that the first identified dengue type in India was DENV4 in Kanpur in 1968, soon after the start of the dengue epidemics, and this was followed subsequently by both DENV2 and DENV4 the following year.
Figure 2: Phylogenetic tree in radiation format of all Indian dengue virus envelope proteins, The four serotypes are clearly separated with DENV1 and DENV3 as the two leaves on the top forming a clade along with DENV2. The DENV4 sequences form a separate clade at the bottom with two sub-clades.
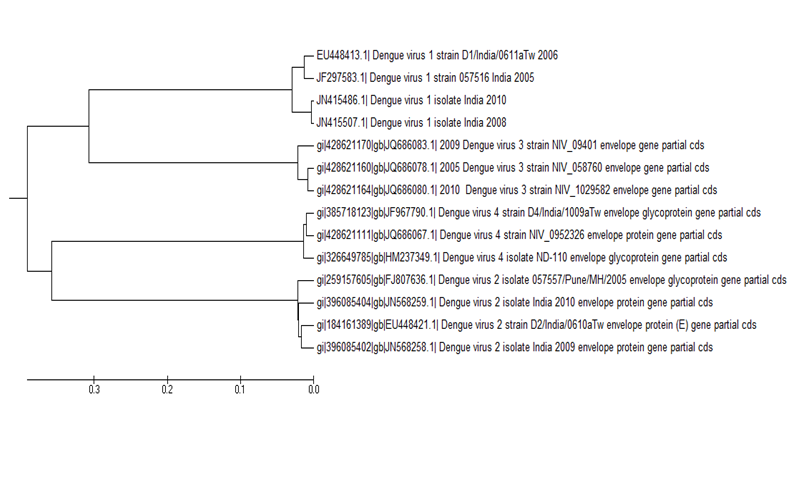
??In another exercise we considered the envelope protein sequences of the four dengue serotypes for recent years, specifically from 2005 onwards, totaling 14 proteins in all, and 19 if we also consider 5 DNV4 sequences from 2012. We had noticed in a dendogram of the envelope genes of all four serotypes of dengue virus of India for the years 2005 to 2010 constructed in an alignment-free model that DENV1,3,4 formed one clade and DENV2 remained in a separate clade distinct from the other three[17]. Redoing the same analysis in an alignment model (MEGA5.2) using the exact same sequences, Figure 3, we found that serotypes 1 and 3 are now clubbed in one clade and serotypes 2 and 4 are clubbed together in another clade. This is quite different from the dendogram of Figure 2 and constitutes an important observation: The root cause lies in the rapid mutational changes that take place in RNA viruses and the differential changes in these dengue sequences result in apparent differences in sequence relationships. We had observed that the DENV4 sequences cluster in two clades, one before 2000 AD and one after. Taking a cue from that, we did another phylogenetic analysis using all envelope sequences prior to 2000 AD; the results (not shown) reproduced the same structure as in Figure 2, and distinct from Figure 3. This substantiates our observation of differential mutations post 2000 AD and implies that without sufficient and adequate number of sequences phylogenetic relationships between hyper variable viral sequences cannot be established with any degree of reproducibility.
Figure 3: Dendogram of 14 envelope protein sequences of all four dengue serotypes from India for the period 2005-2010 constructed in an alignment model (MEGA5.2).
??The mutational changes that characterize sequences such as of dengue viruses are quite distinct from those of, say, mammalian sequences. In a recent work we had contrasted the transition/transversion ratios of Zika genomes[18] with those of mammalian genes. The transition/transversion ratio matrices for the dengue viruses, Table 2, also show similar nature: the transition rates are about 10 times those of transversion rates, whereas for mammalian genes such as beta globins this ratio is around 2 to 3. Such difference was also noticed by[19] in the hyper variable segments of human mitochondrial DNA control region. In case of the dengue viruses too we found that the T-C transition rates are much higher for all serotypes than the A-G transitions; also, the purine to pyrimidene transversion rates is smaller than for the pyrimidene to purine transversion rates. The same effect was noticed in the case of the new Zika virus genomes also[18] and appears to be a property of the flaviviruses.
Table 2(a - d) : Transition/transversion ratio matrices for four Indian dengue serotype genes.
a) Transition/transversion ratios of DENV1 envelope genes
| FromTo | A | T | C | G |
| A | - | 2.1225 | 2.0291 | 9.9696 |
| T | 3.17 | - | 28.4404 | 2.6613 |
| C | 3.17 | 29.749 | - | 2.6613 |
| G | 11.8751 | 2.1225 | 2.0291 | - |
b): Transition/transversion ratios of DENV2 envelope genes.
| FromTo | A | T | C | G |
| A | - | 2.9275 | 2.6706 | 10.9621 |
| T | 4.5639 | - | 22.608 | 3.4862 |
| C | 4.5639 | 24.7824 | - | 3.4862 |
| G | 14.351 | 2.9275 | 2.6706 | - |
c): Transition/transversion ratios of DENV3 envelope genes.
| FromTo | A | T | C | G |
| A | - | 1.7776 | 1.6367 | 13.2384 |
| T | 2.6062 | - | 26.2932 | 2.2427 |
| C | 2.6062 | 28.5582 | - | 2.2427 |
| G | 15.384 | 1.7776 | 1.6367 | - |
d): Transition/transversion ratios of DENV4 envelope genes.
| FromTo | A | T | C | G |
| A | - | 2.7144 | 2.2357 | 12.2114 |
| T | 3.6046 | - | 22.8378 | 3.2184 |
| C | 3.6046 | 27.7277 | - | 3.2184 |
| G | 13.6768 | 2.7144 | 2.2357 | - |
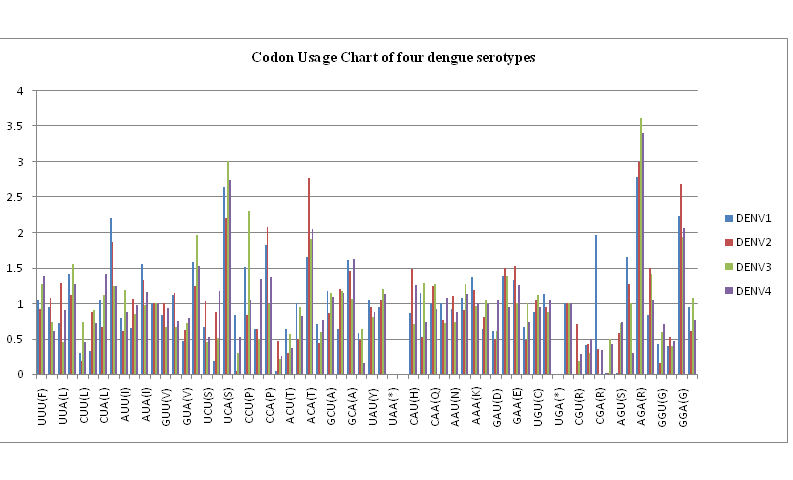
??One of the important parameters in comparative analysis of protein coding nucleotide sequences is the codon usage profiles. Figure 4 shows the relative synonymous codon usage (RSCU) chart for the nucleotide sequences of the envelope genes of the four dengue serotypes in our database of dengue viruses isolated in India. We notice that there are considerable differences between the codon usage preferences between the various serotypes. For example, arginine usage is considerably high for all serotypes, but the codon AGA is exceptionally high for DENV3 and DENV4, whereas arginine by codon CGA is predominant in DENV1. Table 3 lists the details of the codon usage patterns for all the dengue serotypes.
Figure 4: Codon usage bias chart for the envelope protein nucleotide sequences of the four dengue serotypes.
Table 3: Codon usage bias for all the dengue serotypes.
| Codon | DENV1 | DENV2 | DENV3 | DENV4 | Codon | DENV1 | DENV2 | DENV3 | DENV4 |
| UUU(F) | 1.05 | 0.92 | 1.27 | 1.39 | UAU(Y) | 1.05 | 0.95 | 0.8 | 0.87 |
| UUC(F) | 0.95 | 1.08 | 0.73 | 0.61 | UAC(Y) | 0.95 | 1.05 | 1.2 | 1.13 |
| UUA(L) | 0.72 | 1.29 | 0.45 | 0.91 | UAA(*) | 0 | 0 | 0 | 0 |
| UUG(L) | 1.41 | 1.12 | 1.55 | 1.27 | UAG(*) | 0 | 0 | 0 | 0 |
| CUU(L) | 0.3 | 0.18 | 0.74 | 0.45 | CAU(H) | 0.86 | 1.48 | 0.71 | 1.26 |
| CUC(L) | 0.32 | 0.87 | 0.91 | 0.72 | CAC(H) | 1.14 | 0.52 | 1.29 | 0.74 |
| CUA(L) | 1.05 | 0.67 | 1.11 | 1.41 | CAA(Q) | 1 | 1.24 | 1.27 | 0.92 |
| CUG(L) | 2.2 | 1.87 | 1.24 | 1.24 | CAG(Q) | 1 | 0.76 | 0.72 | 1.08 |
| AUU(I) | 0.79 | 0.61 | 1.19 | 0.88 | AAU(N) | 0.92 | 1.1 | 0.73 | 0.87 |
| AUC(I) | 0.65 | 1.06 | 0.85 | 0.97 | AAC(N) | 1.08 | 0.9 | 1.27 | 1.13 |
| AUA(I) | 1.56 | 1.33 | 0.97 | 1.16 | AAA(K) | 1.37 | 1.19 | 0.96 | 1.01 |
| AUG(M) | 1 | 1 | 1 | 1 | AAG(K) | 0.63 | 0.81 | 1.04 | 0.99 |
| GUU(V) | 0.84 | 1 | 0.66 | 0.93 | GAU(D) | 0.61 | 0.5 | 0.61 | 1.05 |
| GUC(V) | 1.11 | 1.15 | 0.66 | 0.75 | GAC(D) | 1.39 | 1.5 | 1.39 | 0.95 |
| GUA(V) | 0.47 | 0.62 | 0.72 | 0.79 | GAA(E) | 1.33 | 1.52 | 1 | 1.26 |
| GUG(V) | 1.58 | 1.24 | 1.96 | 1.53 | GAG(E) | 0.67 | 0.48 | 1 | 0.74 |
| UCU(S) | 0.67 | 1.03 | 0.45 | 0.52 | UGU(C) | 0.87 | 1.05 | 1.12 | 0.95 |
| UCC(S) | 0.18 | 0.87 | 0.51 | 1.18 | UGC(C) | 1.13 | 0.95 | 0.88 | 1.05 |
| UCA(S) | 2.65 | 2.21 | 3.01 | 2.74 | UGA(*) | 0 | 0 | 0 | 0 |
| UCG(S) | 0.84 | 0.04 | 0.29 | 0.52 | UGG(W) | 1 | 1 | 1 | 1 |
| CCU(P) | 1.51 | 0.83 | 2.3 | 1.04 | CGU(R) | 0 | 0.7 | 0.19 | 0.28 |
| CCC(P) | 0.63 | 0.63 | 0.5 | 1.34 | CGC(R) | 0.41 | 0.42 | 0.3 | 0.49 |
| CCA(P) | 1.82 | 2.08 | 0.99 | 1.37 | CGA(R) | 1.97 | 0.36 | 0 | 0.34 |
| CCG(P) | 0.04 | 0.46 | 0.21 | 0.25 | CGG(R) | 0.02 | 0.02 | 0.49 | 0.43 |
| ACU(T) | 0.63 | 0.3 | 0.56 | 0.37 | AGU(S) | 0.02 | 0.58 | 0.72 | 0.74 |
| ACC(T) | 1.01 | 0.5 | 0.94 | 0.82 | AGC(S) | 1.65 | 1.27 | 1.01 | 0.3 |
| ACA(T) | 1.65 | 2.77 | 1.91 | 2.05 | AGA(R) | 2.78 | 2.99 | 3.62 | 3.41 |
| ACG(T) | 0.71 | 0.44 | 0.59 | 0.77 | AGG(R) | 0.83 | 1.5 | 1.41 | 1.05 |
| GCU(A) | 1.18 | 0.86 | 1.14 | 1.09 | GGU(G) | 0.43 | 0.16 | 0.59 | 0.7 |
| GCC(A) | 0.63 | 1.2 | 1.17 | 1.14 | GGC(G) | 0.39 | 0.53 | 0.4 | 0.47 |
| GCA(A) | 1.61 | 1.46 | 1.06 | 1.63 | GGA(G) | 2.24 | 2.69 | 1.94 | 2.07 |
| GCG(A) | 0.58 | 0.48 | 0.63 | 0.15 | GGG(G) | 0.94 | 0.61 | 1.07 | 0.77 |
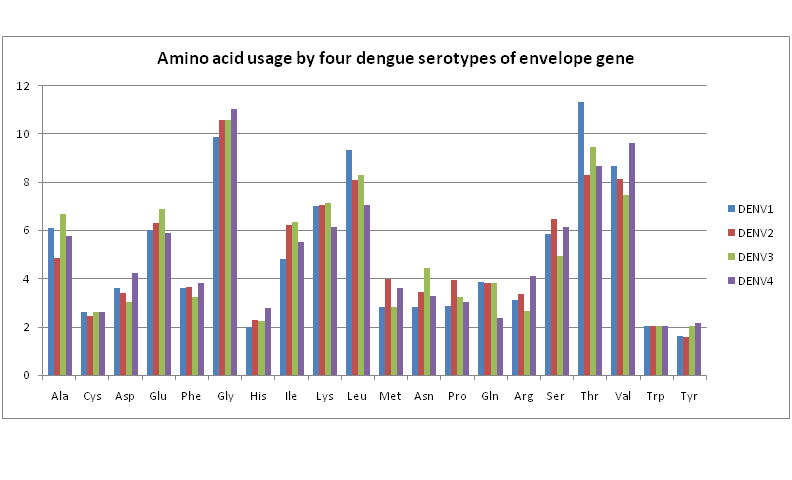
??We therefore next consider amino acid usage in the envelope proteins to determine any significant differences between the four dengue serotypes. Figure 5 shows a bar chart of the amino acid usage patterns. These are almost uniform across all serotypes for all amino acids except for a few noticeable differences: While usage of threonine, and to some extent of leucine also, are rather high in DENV1 compared to the other three serotypes, DENV4 shows quite high usage of valine and some excess of glycine compared to the other serotypes. For all the other amino acids, each serotype seems to have almost the same usage level.
Figure 5: Amino acid usage by four dengue serotypes of envelope gene.
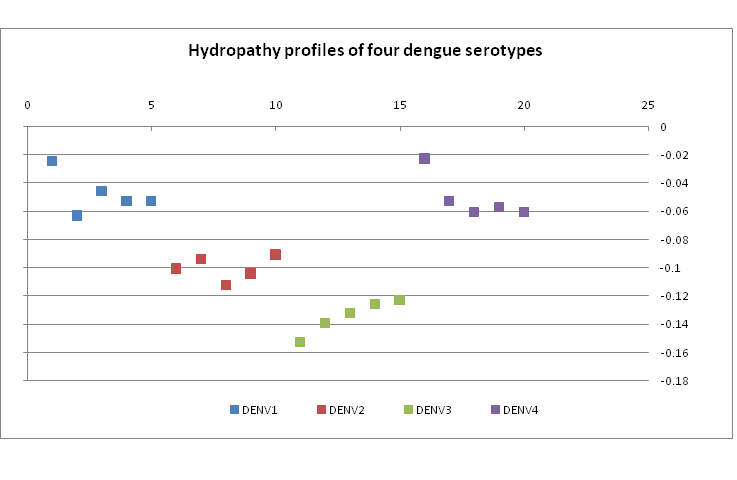
??These observations are borne out by the hydropathy index values of the four dengue serotypes. Figrue 6 shows a plot of five representative samples of hydropathy index of each serotype. DENV3, for example, is seen to be most highly hydropathic. Perusal of the amino acid usage patterns (Figure 5) shows that indeed the usage of such hydrophobic residues as alanine, lysine, isoleucine, etc. is the highest for DENV3. On the other hand, DENV1 and DENV4 have lower hydropathy index indicating more hydrophilic residues; Figure 5 shows that serine, threonine, asparagines, glutamine usage are indeed quite high and generally of comparable percentage.
Figure 6: Hydropathy plot of five examples of each of four dengue serotypes in our database.
??In this context it is instructive to compare and contrast the similarities and dissimilarities of amino acid distribution in the envelope protein sequences. Alignment of the protein sequences of all the four dengue serotypes in our database by MEGA5.2 software shows that 156 residues out of total protein length of 495 residues (for DENV1,2,4; 493 for DENV3) are conserved. Comparing individual serotypes against one another gives various figures show in Table 4 where the absolute number of conserved residues and also those numbers in percentage of the total are shown. It can be seen that DENV1 and DENV3 have more similar segments.
Table 4: Amino acid similarity matrix between four dengue serotypes
| Amino acid similarities | |||
| ? | DENV2 | DENV3 | DENV4 |
| DENV1 | 136 27.47% |
303 61.21% |
236 47.67% |
| DENV2 | ? | 232 46.86% |
208 42.02% |
| DENV3 | ? | ? | 248 50.10% |
??The similarities in structure of the nucleotide sequence as observed from the 2D graphs in Figure1, and the similarities we notice in amino acid usage and hydropathy patterns, suggests that it might be worthwhile to consider what similarities may be observable in the protein structures. Structure of the envelope protein is a dimer having 495 amino acid residues of dengue virus. The envelope dimer of a mature virus particle consists of three domains. Domain I constitute amino acid from 1 - 52, 132 - 191 and 278 - 294, domain II contains amino acid from 53 - 131 and 192 - 277 while domain III consists of amino acids from 295 - 392[14,20]. The rest portion consists the stem segment (amino acids from 394 - 449) and trans membrane anchor portion (amino acids from 449 - 495). The two glycosylation sites are on the 67 and 153 residues of the amino acids. The amino acid regions near the glycosylation site vary among the strains of individual as well as among the serotypes of dengue virus[14,20].
??Alignment of the envelope protein sequences of all four Indian dengue virus serotypes in MEGA5.2 also yields three conserved regions which are listed below Table 5. The first two regions are in the domain II whereas the third region belongs to the stem loop region of the protein.
Table 5: Conserved regions common across all four dengue serotypes from India.
| Regions | Amino acid stretch | Sequence | |
| Start | End | ||
| 1 | 97 | 111 | VDRGWGNGCGLFGKG |
| 2 | 252 | 269 | VLGSQGAMHTALTGATE |
| 3 | 407 | 416 | RKAKRMAILG |
(Bold red letter indicates that the particular amino acid varies in few sequences)
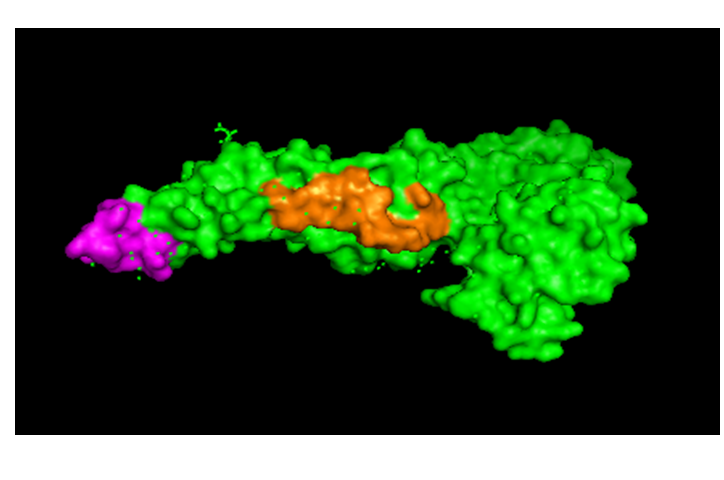
??Downloading the 3-D structure of the envelope protein of the dengue types (3G7T for DENV1 and 1OK8 for DENV2), we located the conserved regions on the protein of DENV2 by using PyMOL. Two of the conserved regions are shown Figure 7 below; the third region could not be indicated since the sequence structure is not included in these structural data. This may be of interest in design of peptide vaccines, work on which is in progress at this time.
Figure 7: Showing the two conserved regions 1 and 2 of envelope protein of DENV2 which are marked in mauve and brown respectively.
Conclusion
??This brief analyses of the main characteristics of the envelope gene and protein sequences of all the four dengue serotypes identified circulating in India and available in the public databases show that genetically the four serotypes have very little differences between them. The base distributions in the gene sequences show strong conserved nature near the 3?-end while phylogenetic analysis have shown that DENV4 is cladistically slightly removed compared to DENV1, DENV3 and DENV2 which form a hierarchy within themselves. The transition/transversion ratios again show little difference between the four serotypes, and the same is true when their amino acid usage patterns are examined. This also led us to identify at least three regions on the dengue envelope protein surface that were well conserved. Thus, while antigenically the four dengue serotypes appear distinct; there is increasing evidence of a continuum of differences diminishing the distinctions between the serotypes[8]. On the other hand we have noticed that apparently differential rates of mutational changes can lead to erroneous relationship indications unless sufficiently large databases are used, where completeness of gene sequences are also to be considered. In view of the fact that the dengue viral structure is found to be related to its pathogenesis[9], these observations carry important consequences for monitoring of dengue viruses for protection of the public and for development of drugs and prophylaxis vaccines.
Conflict of Interest:
??The authors report no conflict of interest.
References
- 1. Whithorn, J., Farrar, J. Dengue. (2010) Br Med Bull 95: 161-173.
- 2. Gubler, D.J. Trends Microbiol 10(2): 100–103.
- 3. Gubler, D.J. Cities spawn epidemic dengue viruses. (2004) Nat Med10 (2): 129–130.
- 4. Anuradha, S., Singh, N.P., Rizvi, S.N., et al. The 1996 outbreak of dengue hemorrhagic fever in Delhi, India. (1998) Southeast Asian J Trop Med Public Health 29(3): 503–506.
- 5. Farrar, J., Focks, D., Gubler, D., et al. Towards a global dengue research agenda. (2007) Trop Med Int Health 12(6): 695-699.
- 6. Gupta, N., Srivastava, S., Jain, A., et al. Dengue in India. (2012) Ind J Med Res136 (3): 373-390.
- 7. Normile, D. Surprising New Dengue Virus Throws a Spanner in Disease Control Efforts. (2013) Science 342(6157): 415.
- 8. Katzelnick, L.C., Fonville, J.M., Gromowski, G.D., et al. Dengue viruses cluster antigenically but not as discrete serotypes. (2015) Science: 349 (6254): 1338-1343.
- 9. Vaughn, D.W., Green, S., Kalayanarooj, S., et al. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. (2000) J Infect Dis 181(1): 2-9.
- 10. Gromowski, G.D., Barrett, N.D., Barrett, A.D.T. Characterization of Dengue Virus Complex-Specific Neutralizing Epitopes on Envelope Protein Domain III of Dengue 2 Virus. (2008) J. Virol 82(17): 8828-8837.
- 11. Nandy, A. A new graphical representation and analysis of DNA sequence structure: Methodology and Application to Globins Genes. (1994) Current Sci 66(4): 309-314.
- 12. Raychaudhury, C., Nandy, A. “Indexing Scheme and Similarity Measures for Macromolecular Sequences”. (1999) Journal of Chemical Information and Computer Science 39(2): 243-247.
- 13. Nandy, A., Nandy, P. On the uniqueness of quantitative DNA difference descriptors in 2D graphical representation models. (2003) ChemPhys Letters 368(1-2): 102-107.
- 14. Modis, Y., Ogata, S., Clements, D., et al. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. (2005) J Virol 79(2): 1223-1231.
- 15. De, A., Sarkar, T., Nandy, A. Bioinformatics studies of Influenza A hemagglutinin sequence data indicate recombination‑like events leading to segment exchanges. (2016) BMC Res Notes 9: 222.
- 16. Ghosh, A., Nandy, A., Nandy, P. Computational analysis and determination of a highly conserved surface exposed segment in H5N1 avian flu and H1N1 swine flu neuraminidase. (2010) BMC Struct Biol 10:6.
- 17. Dey, S., Nandy, A., Nandy,P., et al. Diversity and evolution of the envelope gene of dengue virus type 1 circulating in India in recent times’. (2014) Int. J. Bioinformatics Research and Applications 11(6): 469–482.
- 18. Nandy, A., Dey, S., Basak, S.C., et al. Characterizing the Zika virus genome – A bioinformatics study. (2016) Curr Comput Aided Drug Design 12(2): 87-97.
- 19. Tamara, K., Nei, M. Estimation of the Number of Nucleotide Substitutions in the Control Region of Mitochondrial DNA in Humans and Chimpanzees. (1993) Mol. Biol. Evol 10(3): 512-526.
- 20. Nayak, V., Desau, M., Kucera, K., et al. Crystal structure of dengue virus type 1 envelope protein in the post fusion conformation and its implications for membrane fusio. (2009) J Virol 83(9): 4338-4344.