DNA by Mail: Ensure DNA Integrity by use of Self-Drying Buccal Swabs
René Rohrmanstorfer, Stefanie Zelch, Daniel Wallerstorfer, Sheri Hunt, Chung Hyun, Valentina Kazlova
Affiliation
- 1Amway GmbH, Benzstr. 11 b-c, D-82178 Puchheim, Germany
- 2Novogenia GmbH, Saalachstr. 92, A-5020 Salzburg, Austria
- 3Access Business Group, Buena Park, CA 90621, United States
Corresponding Author
Nicole Bauer, Amway GmbH, Benzstr. 11 b-c, D-82178 Puchheim, Germany E-mail: nicole.bauer@amway.com
Citation
Bauer, N., et al. DNA by Mail: Ensure DNA Integrity by use of Self-Drying Buccal Swabs. (2017) Lett Health Biol Sci 2(2): 71- 77.
Copy rights
© 2017 Bauer, N. This is an Open access article distributed under the terms of Creative Commons Attribution 4.0 International License.
Keywords
DNA Concentration; DNA Stability; Environmental Storage Conditions; PCR Polymerase Chain Reaction; Self-drying Buccal Swabs
Abstract
The goal of this research was to evaluate the quantity and quality of deoxyribonucleic acid (DNA) material collected by at-home-sampling using self-drying buccal swabs in a sterile, laboratory setting compared against a 10,000+ subject sample of real-world swabs processed by consumers and analyzed by a hub-lab of the Sponsor using the same extraction and analysis methodology. Buccal swab DNA concentration was observedon two variables: DNA concentration over time, and DNA sample response to environmental conditions.
Introduction
The collection of buccal cells is a noninvasive, reliable method to obtain genomic deoxyribonucleic acid (DNA) for genetic association studies. The collection may be carried out in-home, by a consumer. One safe, cost-efficient, simplistic, and commonly used technique for home collected DNA samples is the buccal swab technique[1,2].
The processes of DNA collection, handling, and storage of the buccal swab, however, have been identified as critical steps for the overall success rate of the DNA analysis. The swab collection and the handling post collection may be the most determining variables as to whether a sample will maintain its integrity or be rendered useless due to complications such as contamination, degradation, and insufficient yield. As published by Walker et al.[3] the overall success rate for mailed samples decreases to 77.6 % using PCR as opposed to 96.1% for fresh-processed swabs using the same extraction approach.
Several differential factors occur in the process of sample collection given the consumer is a new sample collector. These include, but are not limited to, the time and duration of swabbing, the swab contact location, and the pressure of the swab technique and the drying of the swab after sampling. These added variables from a self-collection technique are considered significant. Feigelson et al[4] could show that tooth is brushing one hour before sample collection reduced the amount of DNA by 40 %.
The success rate of DNA extraction is dependent on both the number of buccal cells collected and the stability of the sample on the swab itself. Microorganism growth during transportation from the consumer´s home to the laboratory should be minimized. Microorganism growth increases with temperature and water activity and reaches optimum incubation potential at +37°C[5]. Given the sub-optimal testing environment, home-collected and self-administered swabs have a lower probability of being completely dry, and an increased probability of contaminant incubation.
Also, cotton or viscose swabs are reported to be conducive to microorganism growth. García-Closas et al[6] studied the quality of home-collected and mailed-in DNA samples, whereby the mailing and room temperatures of the swab location were considered, respectively, with a two-day transit time evaluated. DNA concentration was analyzed based on molecular weight; 55% to 60% of home-collected and room-temperature-mailed buccal swabs were non-degraded, andconsidered to be high molecular weight, with sufficient DNA material, allowing for successful extraction, where 40% to 45% of swabs could not be extracted due degradation of the DNA to low molecular weight fragments. For a successful genetic analysis, the extractable amount of DNA from buccal swabs must meet a minimum amount. Saftlas et al[7] report that the lowest amount of total DNA from a buccal swab to be successfully genotyped is 6 μg.
Due to the moisture wicking potential of a non-viscous swab, there is potential for such a swab to act as asubstrate for the growth of microorganisms. It may be concluded that a rapid decrease of water activity on the swabs will minimize the microorganism growth and consequently,preserve DNA integrity.
Given that a highly stable, well-ventilated swab capable of DNA collection in non-laboratory conditions is needed for at-home administration, this experiment sought to demonstrate that the quality of home-collected and mailed DNA samples may be optimized using self-drying buccal swabs. This specific swab-type benefits from a self-drying process through its ventilation membrane, the consumer is only required to simply place the swab into the drying tube following sample collection.
Objective of this study
To evaluate swab stability, this experiment sought to simulate a stress test of various shipping situations, designed to identify if swabs remained stable over time, and in various environmental conditions. Subsequently, these stress tests were evaluated and benchmarked on real world data from the Sponsor’s hub laboratory; this comparative review of results provide meaningful perspective to the applicability of the stress test in this formalized clinical evaluation. In the formal laboratory environment, three model scenarios simulated the process of postal delivery under different conditions: hot/humid, freeze/thaw, and a most generalized condition, included as the control arm. Each scenario was tested on samples obtained from three adults. Over a fifteen-day study duration, the DNA concentration was quantified using spectrophotometry on day zero and every third day. It was considered that the amount of extractable DNA from swab to swab may be variable, potentially subject dependent, contingent on swabbing technique, and affected by some samples taken from a single subject, the baseline DNA concentration was evaluated for all swabs to ensure substantially equivalent concentration measures across the subjects evaluated samples. This baseline informationwas recorded for reference, subjects were sampled per protocol, and the samples were randomly allocated to the environmental condition experiments.
Additionally, real world data was collected from a large sample following the same methodology to compare results from the formal laboratory setting. A hub lab for the sponsor analyzed the results on 10,653 subjects with all possible conditions and shipping times varying from 3 days to 14 days. Genotype failure rate, defined as the inability to analyze all 5 SNPs in triplicate swabs accurately, was recorded and compared to the laboratory data.
Materials and Methods
Formal Laboratory Setting
Self-drying Buccal Swabs (Sarstedt™)
Self-drying buccal swabs from Sarstedt GmbH / Germany (forensic swab no. 80.629) were used to collect DNA samples. These swabs are equipped with a ventilation membrane at the tube base, which enables self-drying of the swab in the tube. Samples may be put into the tube immediately after collection and no drying of the swab in the open, outside the tube, is required. Furthermore, due to the rapid self-drying process, using these types of swabs is expected to reduce the risk of DNA contamination and degradation forhome-collected, mailed DNA samples.
Subjects and Sampling
Buccal swabs samples were collected from three adults (n = 3) at Novogenia GmbH, Salzburg, Austria. For each subject, three sets of 18 swabs were collected during three different swabbing days to accommodate swab stability in the three environmental conditions (total swabs for environment experiment = 162). Also, 18 other swabs from each of the three subjects were collected and analyzed separately to evaluate if the DNA yield differed with a consecutive collection of swabs between the 1st and 18th swab (total swabs for DNA concentration in consecutive samplings = 54). This analysis was needed to substantiate the randomization of swabs that took place for the evaluation of environmental impact on swab stability.
Subjects confirmed that proper pre-study requirements had been met, including not having brushed their teeth in the last 3 hours and not having eaten within 3 hours, consuming only water for the prior 3 hours. Site technician administered swabs, 18 per day for three consecutive days, to the inside of each of the cheeks by rubbing over the lower gap between the gumline and cheek. The technician wore a protective mask and eyewear and appropriate gloves to prevent contamination of the swabs and subject.
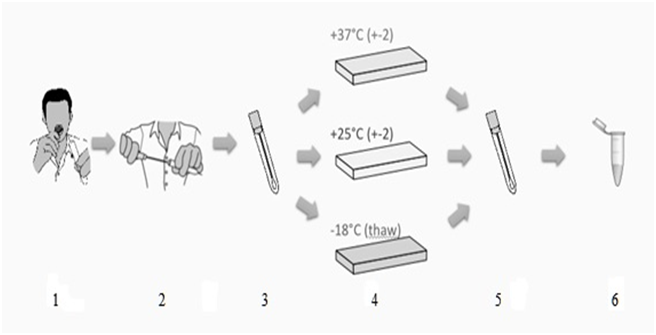
For each topic, a total of 72 swabs were collected (4 sets of 18 swabs). One set from each subject was used to study the baseline variability of extractable DNA. The three other sets (54 swabs per subject) were randomized into six boxes, each containing three different swabs. These boxes were then randomly allocated to three experiments and analyzed at day 0, 3, 6, 9, 12 and 15 (total number of swabs collected = 216). Table I provides an overview on the samples collected. Figure 1 explains the sampling procedure.
Figure 1: Process flow Swab handling:
1) Swabs were taken for 30 s each. No food or beverage has been consumed for at least 3 h before sampling.
2) Swabs were returned to the self-drying tube and sealed.
3) Swabs were transferred into a cardboard box and boxes were stored at 3 different environmental conditions.
4) Swabs were taken from these conditions at various intervals for further processing.
5) DNA was extracted from swabs and resulting DNA concentration was measured.
Table I: Total swabs analyzed: formal laboratory setting.
| Experiment | Number of Swabs |
|---|---|
| Baseline Variability | 54 |
| Environmental Variability | 162 |
| Total Swabs Analyzed | 216 |
DNA Extraction
DNA was extracted using the QIA cube robot with silica membrane spin column technology (QIAGEN Blood Mini Kit). Buccal swabs were prepared by incubation in 150 μL acetate lysis buffer (kit content) and 30 μL proteinase K for 15 minutes at 56°C.
The tube was vortexed for 10s and centrifuged for 120 s at 3200 rpm. The swab was removed while leaving the main fraction of lysate in the tube. The lysate was then loaded into the QIA cube robot and extracted using the blood mini kit automation protocolwith QIA amp Mini SpinColumns. The sample was loaded onto the column and purified using ethanol and AW1 and AW2 wash buffers. The final DNA sample was eluted in 100 μL TE buffer.
UV-Vis Spectrophotometry
The concentration of DNA extractable materials was quantified using UV-Vis Spectrophotometry. The absorption of UV light by DNA provides a straight forward and accurate estimation of the concentration of nucleic acids in a sample. This standardized approach was used on day zero, and every third day following the swabbing of the individuals. This method is accurate within one ng/μL as described in the NanoDrop 1000 Spectrophotometer V3.7 User’s Manual[8].
Process summary
Experiment conditions: Temperature and Humidity: The experiment was conducted simulating three postal delivery scenarios to evaluate swab stability in conditions that are likely to be experienced during shipping: hot/humid, freeze/thaw, and optimum conditions. Within a period of 15 days, every 3rd day the extractable DNA from swabs was quantified using spectrophotometry. (Table II)
Table II: Temperature and humidity conditions per experiment.
| Temperature | Relative Humidity | ||
|---|---|---|---|
| Experiment 1 | Optimum | +25 ± 2°C | ≤ 50% |
| Experiment 2 | Freeze/thaw | -18 ± 2°C | Not Applicable |
| Experiment 3 | Hot/Humid | +37 ± 2°C | 80 ± 10% |
| Baseline Variability | Optimum | +25 ± 2°C | ≤ 50 % |
Data Analysis: Statistical variance analysis (ANOVA) was conducted using the statistical analysis software JMP (SAS Institute). Threshold concentration was set to 10 ng/μL, and concentration values below this were considered as not containing sufficient amounts to be successfully extracted and analyzed. Swabs were all analyzed regardless of the DNA concentration failure per 10 ng/μL thresholds. ‘Genotype failure’ refers to the inability to correctly genotyped the swab.
Parameters observed and calculated were the following:
1. The overall distribution of DNA concentration values was computed; overall mean and standard deviation was calculated.
2. For each subject, mean and standard deviation of DNA concentration values were calculated separately.
3. Number and percentage of failures (both DNA Concentration and Genotype failure) were evaluated across different temperature/humidity conditions, as well as days, replica number, subject ID.
4. ANOVA was conducted using two-way repeated measures.
Materials and Methods-Real World Laboratory: 10,653 individuals were supplied with 3 self-drying buccal swabs (Sarstedt™) and instructions to collect the sample by rubbing the swab across the inside of the mouth in various positions (including gutter, cheek, tongue, and palate) for 30 s each. Collection time was instructed to be the first thing in the morning before eating, drinking or brushing teeth. These swabs were then shipped via courier in cardboard boxes identical to the boxes used in the stability experiment. Shipment times varied from 5 to 14 days and conditions were not measured. However, the normalized variance is expected in these real-world conditions, as the formalized experiment sought to mirror duration and temperature/humidity fluctuations specific to shipments subject to European countries’ range of shipment time and seasonal temperature fluctuations.
Swabs were unpacked upon arrival and stored at 20 - 25°C and relative humidity of approximately 20 - 30%. For each, the first swab was extracted, and 5 SNPs were analyzed according to the same methodology as in the laboratory settings to determine success rates in genotyping the sample. If at least one of the 5 SNPs yielded a low or no signal, the second swab was extracted and analyzed for all 5 SNPs. If the result of the second swab was combined with the result of the first swab and at least one SNP still failed to be genotyped, the third swab was extracted and analyzed. If the combination of all 3 results still yielded gaps of at least 1 SNP, the sample was evaluated to be failed.
Results
Baseline Variability
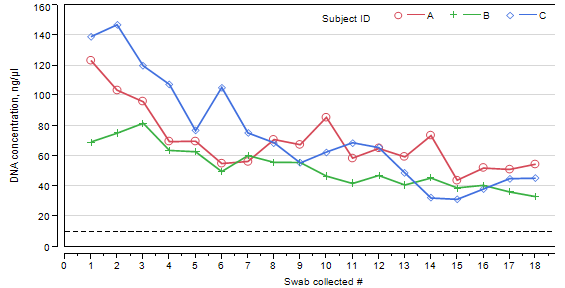
By conducting two-way ANOVA, it was determined that the amount of DNA differ significantly between subjects (p-value = 0.0003) and declines significantly with the increased number of consecutive swabs (p-value < 0.0001). However, when swabbed 18 times, the amount of extractable DNA material is still above a threshold concentration of 10 ng/μL at optimal conditions (Figure 2, Table IV). In the “at home” environment, only three swabs are collective consecutively. Results of repeated measures ANOVA showed that in the first three swabs the swab number does not have a significant effect on the amount of DNA extracted (p-value = 0.5509).
Figure 2: DNA concentration across consecutive swabs. Each line is represented by observations from a separate subject. Dashed line on the bottom of the plot is corresponding to the threshold of 10 ng/μL.
Environmental Variability
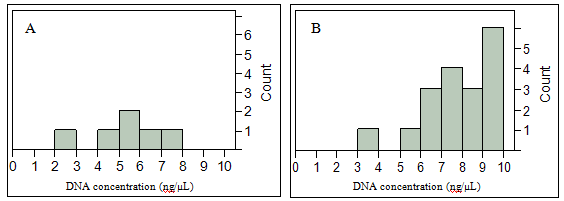
A total of 162 swabs from three subjects in the Formal Laboratory Setting were analyzed to evaluate the impact of time and storing conditions (Table III). 24 swabs failed with the DNA concentration below 10 ng/μL (14.8% of total swabs), and the average amount of DNA material was 29.84 ng/μL with a standard deviation of 28.04 ng/μL. For each subject, a total of 54 swabs were submitted to different temperature and humidity conditions. Out of 162 swabs in the experiment, 14.8 % had the amount of DNA below suggested threshold (10 ng/μL). However, only 3.7 % of swabs failed to be genotyped (25% of those that fell below the threshold). Samples with the DNA concentration higher than 7.6 ng/μL were all genotyped successfully. Samples that had concentration 7.6 ng/μL and lower were still genotyped successfully at a rate of 57 % (Figure 3, Table V-VII).
Table III: Environmental variability in analytical threshold.
| Group | Label | N | N with DNA concentration ≤10 ng/μL | % with DNA concentration ≤10 ng/μL | N failed to be genotyped | % failed to be genotyped |
|---|---|---|---|---|---|---|
| Swabs total | n/a | 162 | 24 | 14.8 | 6 | 3.7 |
| Person | A | 54 | 5 | 9.3 | 0 | 0 |
| B | 54 | 2 | 3.7 | 0 | 0 | |
| C | 54 | 17 | 31.5 | 6 | 11.1 | |
| Temperature / Humidity conditions | Freeze/Thaw | 54 | 2 | 3.7 | 0 | 0 |
| Optimum | 54 | 8 | 14.8 | 0 | 0 | |
| Hot/Humid | 54 | 14 | 25.9 | 6 | 11.1 | |
| Day | 0 | 27 | 4 | 14.8 | 0 | 0 |
| 3 | 27 | 5 | 18.5 | 1 | 3.7 | |
| 6 | 27 | 6 | 27.2 | 1 | 3.7 | |
| 9 | 27 | 1 | 3.7 | 1 | 3.7 | |
| 12 | 27 | 2 | 18.5 | 3 | 11.1 | |
| 15 | 27 | 3 | 11.1 | 0 | 0 |
Figure 3: Distributions of DNA concentration for swabs that fell below the threshold of 10 ng/μL: A = analysis failed, B = analysis successful
Furthermore, all swabs that failed to be genotyped were part of the high temperature/high humidity condition. Contextually, one of the samples with a DNA concentration as low as 3.8 ng/μL that came from freeze/thaw condition was still genotyped successfully. Based on the figure above, there is a trend toward higher temperature and humidity conditions being associated with more failures in genotyping.
Observations related to temperature / humidity conditions
Repeated measures ANOVA was conducted to assess the number of samples that failed below the threshold (10 ng/μL) across experimental conditions (obtained p-value = 0.2066)[9].
There is insufficient evidence at the α = 0.05 level of significance to reject the hypothesis that there is a difference in DNA concentration when storing buccal swabs at normal, cold or hot conditions. In other words, temperature/humidity conditions do not have a significant effect on swab stability.
However, a clear trend in failures was observed with the hot/humid condition. While not statistically significant to predict failures, this qualitative observation does give perspective to several of the samples that fell below the 10 ng/μL threshold that also failed to genotype (Figure 3).
Real World Laboratory Data
After analyzing 10,653 subject shipments from the Sponsor’s hub-laboratory, a total of 10,582 subjects were genotyped successfully, while 71 subjects failed on each of the three swabs included in the consignment to the laboratory, resulting in a failure rate of 1 in 150 subjects or 0.7 %.
While the real-world laboratory results show a subject failure rate of 0.7%, the actual swab failure rate would be higher, given consumers send in samples in triplicate, where a subject’s failure is only noted when all three swabs cannot be genotyped. To compare real, recorded data, we accept the distinction that the real-world experiment considered subject failures, while the formal laboratory experiment examined swab failures.
Discussion
According to published science[10] using standard buccal swabs, the amount of extractable DNA is variable and, among other aspects, dependent on the subject that provides the material. When quantifying the amount of DNA on consecutively collected samples, the biggest variation from one swab to another was linked to the subject. Among the three subjects, the rate of failure below 10 ng/μL varied from 3.7% for Subject B to 31.5 % for subject C. All swabs that failed to be genotypedwere collected from subject C.
Also, subjects were swabbed 18 times consecutively to evaluate the baseline variability in a stress laboratory setting. For all swabs the amount of extractable DNA material was still above a threshold concentration of 10 ng/μL at optimal conditions but using the two-way ANOVA, it was determined that the amount of DNA collect declines significantly when swabbing subjects consecutively. Therefore, we can qualitatively assess that DNA concentration will drop after numerous, consecutive sampling takes place. In the at home environment, subjects selfswab in triplicate, avoiding the potential of tapered DNA concentration from swab-to-swab due to consecutive sampling. Results of repeated measures ANOVA showed that in the first three swabs the swab number does not have a significant effect on the amount of DNA extracted (p-value = 0.5509).
Based on the 14.8% failure rate across all conditions and timelines (DNA yield below 10 ng/μL), a rate of 14.8% samples would fail to meet minimum criteria. Including 2 swabs in a sample kit would decrease the failure rate to (0.148 * 0.148 = 0.0219) 2.2 %, meaning that 1 in 45 subjects would have to be recalled. Adding a third swab reduces the failure rate to (0.148 * 0.148 * 0.148 = 0.00324) 0.32%, meaning only one in 312 subjects need to be recalled.
Using the established system with the same methodology in a real-world environment, a hub lab for the Sponsor analyzed its results on 10,653 subjects with all possible conditions and shipping times varying from 3 days to 14 days. The analysis of this laboratory’s internal data showed that the use of three swabs leads to a historical recall rate of 71 in 10,653 subjects, which equates to a failure rate of 1 in 150 or 0.7%. Given the large-scale, real-world conditions show a higher failure rate, attributable to a variety of variables created by a consumer self-swabbing, the real-world data evaluation and outcomes support the validity of the Formal Laboratory Setting’s results explained herein, executed with a smaller number of subjects.
It is important to note that besides DNA quantity, as this study sought primarily to evaluate, DNA quality is also a crucial factor as the polymerase chain reaction (PCR) technology can compensate for low DNA yields by amplification if sufficient high quality starting material is available. In DNA samples collected at home, it is known that DNA quality typically declines with DNA yield[11]. The present study limited its focus to DNA yield as a measure of overall sample quality, but DNA quality must not be ignored in this respect. In a simple attempt to gain insight into the quality aspect of DNA in low quantity samples, genotyping experiments for 5 established SNP genotyping assays were performed on each sample below 10 ng/μL. Only 25 % of the samples below the threshold of 10 ng/μL failed to be genotyped due to insufficient acceptable quality DNA. While notable, but not significant, these samples came from the Hot/ Humid condition. This suggests that the hot and humid condition may influence DNA quality, which is, however, not sufficient to cause significant failure numbers at 14 days in transit.
Conclusion
Collecting DNA samples using self-drying buccal swabs (Sarstedt™) at home is a reliable method to obtain quality samples for DNA genotyping. Because of the self-drying process created by these swabs’ design, contamination potential is reduced, and the amount of DNA material and quality is stable over time and under various environmental conditions.
While the Formal Laboratory Experiment evaluated a small subject sample, this analyzed data benefits from substantiation by a massive, real-world laboratory sample which corroborates the conclusion of overall stability of DNA collected on self-drying buccal swabs (Sarstedt™), even when subjected to a stress test of environmental conditions that may be experienced during transit from the at-home environment to a genetic laboratory.
Acknowledge:
The authors would like to thank Dan Brenner and Marine Herrenknecht for their help with the compilation of the manuscript. (SPRIM Strategy & Intelligent Innovation, Orlando, FL 32826, United States). Additional thanks to Sarstedt GmbH Company for the free donation of Sarstedt™ swabs for this study.
References
- 1. Le Marchand, L., Lum-Jones, A., Saltzman, B., et al. Feasibility of collecting buccal cell DNA by mail in a cohort study. (2001) Cancer Epidemiol Biomarkers Prev 10(6): 701-703.
Pubmed || Crossref || Others - 2. Freeman, B., Powell, J., Ball, D., et al. DNA by mail: an inexpensive and non-invasive method for collecting DNA samples from widely dispersed populations. (1997) Behav Genet 27(3): 251-257.
Pubmed || Crossref || Others - 3. Walker, A.H., Najarian, D., White, D.L., et al. Collection of genomic DNA by buccal swabs for polymerase chain reaction-based biomarker assays. (1999) Environ Health Perspect 107(7): 517-520.
Pubmed || Crossref || Others - 4. Feigelson, H.S., Rodriguez, C., Robertson, A.S., et al. Determinants of DNA yield and quality from buccal cell samples collected with mouthwash. (2001) Cancer Epidemiol Biomarkers Prev 10(9): 1005-1008.
Pubmed || Crossref || Others - 5. Petersen, J.W. Bacterial Pathogenesis. (1996) Medical Microbiology.
Pubmed || Crossref || Others - 6. Garcia-Closas, M., Egan, K.M., Abruzzo, J., et al. Collection of genomic DNA from adults in epidemiological studies by buccal cytobrush and mouthwash. (2001) Cancer Epidemiol Biomarkers Prev 10(6): 687-696.
Pubmed || Crossref || Others - 7. Saftlas, A.F., Waldschmidt, M., Logsden-Sackett, N., et al. Optimizing buccal cell DNA yields in mothers and infants for human leukocyte antigen genotyping. (2004) Am J Epidemiol 160(1): 77-84.
Pubmed || Crossref || Others - 8. Nano Drop ND-1000 Spectrophotometer User´s Manual. Thermo Fisher Scientific 2008.
Pubmed || Crossref || Others - 9. Schmaal, L., Veltman, D.J., Nederveen, A., et al. N-acetylcysteine normalizes glutamate levels in cocaine-dependent patients: a randomized crossover magnetic resonance spectroscopy study. (2012) Neuro psychopharmacology 37(9): 2143-2152.
Pubmed || Crossref || Others - 10. Caboux, E., Lallemand, C., Ferro, G., et al. Sources of pre-analytical variations in yield of DNA extracted from blood samples: analysis of 50,000 DNA samples in EPIC. (2012) PLoS One 7(7): e39821.
Pubmed || Crossref || Others - 11. Looi, M.L., Zakaria, H., Osman, J., et al. Quantity and quality assessment of DNA extracted from saliva and blood. (2012) Clin Lab 58(3-4): 307-12.
Pubmed || Crossref || Others