Genetic Diversity Analysis of Six Major Cereal Crops Cultivars through RAPD Markers
Rupal Chauhan1*,Yogesh Jasrai1, Himanshu Pandya1, Raman Gami2, Kapil Tiwari2
Affiliation
1Applied Botany Center, Department of Botany, University School of Sciences, Gujarat University, Ahmadabad Gujarat, India
2Department of Genetics & Plant Breeding, C.P. College of Agriculture, S.D. Agricultural University, Dantiwada, Gujarat, India
Corresponding Author
Chauhan, R. Applied Botany Center, Department of Botany, University School of Sciences, Gujarat University, Ahmadabad 380 009, Gujarat, India. E-mail: chauhanrupal757@gmail.com
Citation
Chauhan, R., et al. Genetic Diversity Analysis of Six Major Cereal Crops Cultivars through RAPD Markers. (2015) Bioinfo Proteom Img Anal 1(1): 20- 24.
Copy rights
© 2015 Chauhan, R. This is an Open access article distributed under the terms of Creative Commons Attribution 4.0 International License.
Keywords
Genetic Diversity; RAPD Markers; Six major cereal crops (rice, wheat, maize, barley, pearl millet and sorghum); Improved cultivars.
Abstract
The aim of this study was to evaluate the genetic diversity of Indian improved six major cereal crops like rice, wheat, maize, barley, pearl millet and sorghum varieties. The diversity of thirty-six varieties of six major cereal crops (rice, wheat, maize, barley, pearl millet and sorghum) was estimated by Random Amplified Polymorphic DNA (RAPD) technique using 20 primers. RAPD analysis detected a total of 168 DNA fragments of which 107 (63.69%) were polymorphic. The genetic similarity matrix (based on Dice index) ranged from 0.435 to 0.81. These indices were used to construct a dendrogram using average linkage between groups. Results illustrate the potential of RAPD markers showed considerable potential for estimating genetic diversity among our six major cereal crops cultivars. The study provides valuable information for cultivars fingerprinting at a molecular level and can be used to assist efficient selection in six major cereal crops breeding strategies aiming at achieving sustainability in six major cereal crops production in India.
Introduction
The development of DNA marker technology has provided an efficient tool to facilitate plant genetic resource conservation and management. Compared to morphological analysis, molecular markers can reveal differences among accessions at DNA level. They represent an opportunity to provide information on the variation that exists in a particular species within a local region as well as among different countries. They serve as a valuable guide for effective collection and use of genetic resources too. Molecular markers provide information that helps in deciding the distinctiveness of species and their ranking according to the number of close relatives and phylogenetic position[1]. Several types of molecular markers are available for evaluating the extent of genetic variation in rice, wheat, maize, barley, pearl millet and sorghum. These include RFLP[2,3], RAPD[4], AFLP[5] and SSR[6]. Of these RAPD markers is increasingly being employed in genetic research owing to its speedy process and simplicity[2]. This technique always allows the examination of genomic variation without prior knowledge of DNA sequences[7] and is especially useful for unzipping the variations in species with low genetic variability when other techniques such as isozyme analysis fail to reveal differences among the individuals. Moreover, varietal distinctiveness and relativeness can unambiguously be estimated by RAPD fingerprinting in commercially important crops[8]. RAPD markers are considered to be unbiased and neutral markers for genetic mapping applications[9], in population genetics[10], taxonomy[11] as well as for genetic diagnostics. RAPD has been used for classification and assessing diversity and relatedness of six major cereal crops genotypes by several groups[12-17]. This study aimed to use RAPD markers to evaluate the genetic variation within a collection of improved six major cereal crops like rice, wheat, maize, barley, pearl millet and sorghum varieties[18] and to reveal genetic relationships among them for future use in selection, hybridization, biodiversity assessment and conservation of diverse gene pools.
Materials and Methods
Plant Material
emsp; The plant material for the present study comprised of 36 genotypes of six major cereal crops: rice, wheat, maize, barley, pearl millet and sorghum varieties obtained from world collection depository of six major cereal crops at India is given in Table 1. These accessions were maintained at the Botanical Garden of Department of Genetics and Plant Breeding, College of Agriculture, Dantiwada, Gujarat.
Table 1: Six major cereal crops (rice, wheat, maize, barley, pearl millet and sorghum)
| No. | Cereal Crops | Varieties |
|---|---|---|
| 1. | Rice | GC-1 |
| GC-2 | ||
| GC-3 | ||
| GC-4 | ||
| GC-5 | ||
| GC-6 | ||
| 2. | Wheat | GW-11 |
| GW-173 | ||
| GW-496 | ||
| GW-322 | ||
| GW-273 | ||
| GW-503 | ||
| GYL-12 | ||
| GYL-4 | ||
| VL-109150 | ||
| 3. | Maize | VL-109507 |
| HY-207 | ||
| VL-1017749 | ||
| 4. | Barley | RD-2052 |
| BH-933 | ||
| RD-2715 | ||
| RD-2035 | ||
| RD-2552 | ||
| RD-2784 | ||
| 5. | Pearl Millet | GP-38 |
| GP-48 | ||
| GHB-558 | ||
| GP-16 | ||
| GP-49 | ||
| GP-1 | ||
| 6. | Sorghum | CSV-21(F) |
| GJ-39 | ||
| CSV-15 | ||
| GJ-40 | ||
| GFS-5 | ||
| GFS-4 |
Genomic DNA Extraction and Quantification
For isolation of genomic DNA the seeds from six major cereal crops: rice, wheat, maize, barley, pearl millet and sorghum were surface sterilized and sowed in pots containing 1 kg of soil maintained in a greenhouse. Fresh young leaves from 6 plants per cultivar type were collected after 10 day germination and stored at -70°C until further use. Genomic DNA from fresh leaves was extracted by grinding 1g of young leaf sample in liquid nitrogen using chilled pestle and mortar following the procedure of Doyle and Doyle[19] protocol. CTAB extraction buffer [ (N-Iauryl sarcosine, cetyl trimethyl ammonium bromide), 0.2% Mercapto ethanol, 1M Tris-HCl : pH 8.0, 0.1M EDTA :pH 8.0, 1.5M NaCI ], which was pre-warmed to 65°C and added to each tube and placed on a water bath (65°C) for 15 minutes and cooled to room temperature.
Equal volume of chloroform: ‘isoamylalcohol (24: 1) was added and vortexed for few seconds and centrifuged at 13,000 rpm for 10-15 minutes. The supernatant was transferred to a new eppendorf tube. The DNA was precipitated by adding equal volume of prechilled isopropanol and incubated overnight at -20° C. DNA was recovered by centrifugation and the pellet was washed with 70 percent ethanol and dissolved in TE (10mM TrisHCI, 1mM EDTA, pH 8.0) and stored at -20°C. DNA concentration was measured by using Nano Drop and the quality was checked on 0.8% agarose gel by electrophoresis at 100V for 30 min. The gel was stained with ethidium bromide visualized under UV light. RNase 5 μl solution (10 mg ml-1) treatment was given to remove RNA from the samples.
PCR with Random Primers
For RAPD analysis DNA extracted from young leaves from each six major cereal crops cultivar genotype was used as template in the PCR mix. Other components of the reaction mixtures (15 μL total volume) were: 1.5 μl of 10 x PCR buffer, 0.9 μl of 10 mM MgCl2, 1.0 μl of 25 mM dNTPs, 1.5 μl of 10 μM primer, 1.5 μl of template DNA, and 0.2 μl of 1 unit Taq DNA polymerase. DNA Molecular Weight Marker (100 bp ladder; Roche) was used to estimate PCR fragment size. DNA amplification was carried out according to the method of Williams[4]. A total of 20 primers were used for screening in the present study (Table 2). DNA amplification was performed in DNA Thermal Cycler (Eppendorf) programmed as follows: an initial denaturation of 5 min at 94°C; 44 cycles of 94°C for 1 min (denaturation), 37°C for 1 min (annealing), and 72°C for 2 min (extension). One additional cycle of 8 min at 72°C was used for final extension. The PCR amplified products were resolved on 1.5% agarose gel electrophoresis at 72 V for 2 hours and visualized by ethidium bromide staining and the Gel was photographed in Gel Documentation System (VilberLourmat).
Table 2: Number of bands amplified per primer, number and percentage of polymorphism in six major cereal crops (rice, wheat, maize, barley, pearl millet and sorghum) genotypes.
| No. | Primers | Sequences | Total No. of bands | No. of polymorphic bands | % polymorphism |
|---|---|---|---|---|---|
| 1 | OPA-8 | 5'GTGACGTAGG | 11 | 8 | 72.7 |
| 2 | OPA-11 | 5' CAATCGCCGT | 8 | 7 | 87.5 |
| 3 | OPB-7 | 5'GGTGACGCAG | 10 | 7 | 70 |
| 4 | OPB-12 | 5'CCTTGACGCA | 8 | 4 | 50 |
| 5 | OPB-20 | 5'GGACCCTTAC | 10 | 3 | 30 |
| 6 | OPC-6 | 5'GAACGGACTA | 10 | 8 | 80 |
| 7 | OPC-11 | 5'AAAGCTGCGG | 7 | 5 | 71.4 |
| 8 | OPC-14 | 5'TGCGTGCTTG | 10 | 9 | 90 |
| 9 | OPP-4 | 5'GTGTCTCAGG | 10 | 7 | 70 |
| 10 | OPP-8 | 5'ACATCGCCCA | 13 | 11 | 84.6 |
| 11 | OPP-9 | 5'GTGGTCCCCA | 13 | 9 | 69 |
| 12 | OPP-10 | 5'TCCCGCCTAC | 7 | 3 | 42.8 |
| 13 | OPP-11 | 5'AACGCGTCGG | 9 | 7 | 77 |
| 14 | OPP-13 | 5'GGAGTGCCTC | 11 | 8 | 72.7 |
| 15 | OPP-14 | 5'CCAGCCGAAC | 10 | 3 | 30 |
| 16 | OPP-01 | 5'GTAGCACTCC | 11 | 7 | 63.6 |
| 17 | OPA-14 | 5'TCTGTGCTGG | 0 | 0 | 0 |
| 18 | OPP-16 | 5'CCAAGCTGCC | 10 | 1 | 10 |
| 19 | OPC-04 | 5'CCGCATCTAC | 0 | 0 | 0 |
| 20 | OPA-09 | 5'GGGTAACGCC | 0 | 0 | 0 |
| Total | 168 | 107 | 63.69 |
RAPD Product Scoring and Data Analysis
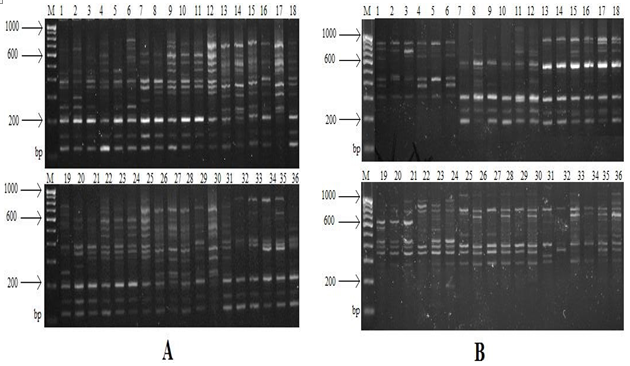
The molecular weight of RAPD polymorphism between six major cereal crops genotypes all bands was calculated by comparison with the 100 base pair marker (Figure 1). All reactions were repeated at least twice and only reproducible bands were scored for statistical analysis. Other visible bands were not considered because of their ambiguous nature. The presence of band taken as ‘1’ or absence of band taken as ‘0’. Estimates of genetic similarity (F) were calculated between all pairs of the cultivars according to[20] based on following formula: Similarity (F) = 2Nab/ (Na + Nb) Where Na = the total number of fragments detected in individual ‘a’; Nb = the total number of fragments shown by individual ‘b’ and Nab = the number of fragments shared by individuals ‘a’ and ‘b’. The resulting similarity coefficients were used to evaluate the relationships among cultivars with a cluster analysis using an unweighted pair group method with arithmetic averages (UPGMA). The analysis was plotted in the form of a dendrogram. All computations were carried out using the NTSYS-pc (numerical taxonomy and multivariate analysis system program), Version 2.2 package[21].
Figure 1: RAPD profiles of thirty-sic different cultivars of six major cereal crops (rice, wheat, maize, barley, pearl millet and sorghum) using 20 primers here only shown two primer result OPC-14 (A) and OPP-16 (B) M: Molecular weight marker (100 bp DNA ladder), Lines: (Pearl Millet)1: 'GP-38', 2:' GP-48', 3:' GHB-558', 4:' GP-16', 5:' GP-49', 6:' GP-1'; (Maize)7:' GYL-12', 8:' GYL-4', 9:' VL-109150', 10:' VL109507', 11:' HY-207', 12:' VL-1017749'; (Sorghum)13:' CSV-21(F)', 14:' GJ-39', 15:' CSV-15', 16:' GJ-40', 17:' GFS-5', 18:' GFS-4'; (Barley)19:' RD-2052', 20:' BH-933', 21:' RD- 2715', 22:' RD-2035', 23:' 'RD-2552', 24:' 'RD-2784'; (Wheat)25:' GW-11', 26:' GW173', 27:' GW-496', 28:' GW-322', 29:' GW-273', 30:' GW-503'; (Rice)31:' GC-1', 32:' GC-2', 33:' GC-3', 34:' GC-4', 35:' GC-5', 36:' GC-6'.
Results and Discussion
In this study 168 DNA fragments reproducible and scorable amplification products were generated across 36 cultivars and these were in the size range of 350 to 1800 bp with an average of 4.6 alleles per cultivar. Out of 168 bands, 107 (63.69%) (Table 2) were found to be polymorphic for one or more cultivars ranging from 5.35 polymorphic bands per primer. The average number of polymorphic fragments per primer among the 36 cultivars of six major cereal crops like rice, wheat, maize, barley, pearl millet and sorghum was 5.35. This percentage of polymorphic bands was similar to that observed in the study[13-17]. It is generally reported that polymorphism between cultivars can arise through nucleotide changes that prevent amplification by introducing a mismatch at one priming site; deletion of a priming site; insertions that render priming site too distant to support amplification and insertions or deletions that change the size of the amplified product[22]. This discrepancy may relate to genotypes and the selection of RAPD primers with scorable bands. The other reason could be the use of more diverse genotypes. In this study, 20 RAPD primers were examined and 16 of them generated scorable bands. In addition, investigated genotypes in this study were chosen from improved cultivars and not from diverse gene pools.
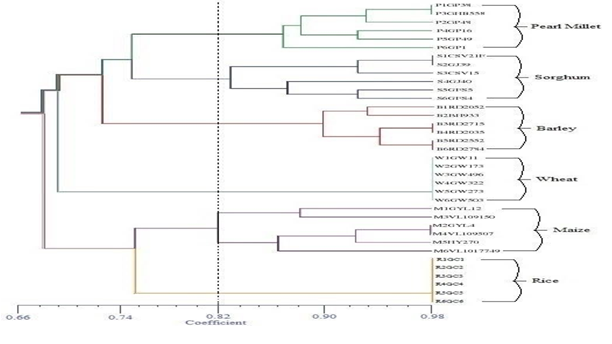
Primers OPC-14 and OPA-11 gave the highest percentage of polymorphic bands, while the minimum polymorphism was observed using OPP-16 primer. A similarity matrix based on the proportion of shared RAPD fragments was used to establish the level of relatedness between improved cultivars. The genetic similarity (GS) for pairs of cultivars was calculated using pair wise Jaccard’s coefficients for the genetic similarities among the 36, based on the data of 20 RAPD primers, were used to construct a dendrogram (Figure 2) using Average Linkage (Between Groups) and SPSS computer program. The similarity indices based on all possible pairs of cultivars ranged from 0.435 to 0.81. The lowest value of pair-wise similarity index was between Pearl Millet (GP-49) and Rice (E-39) cultivars (GS = 0.435).
Figure 2: Dendrogram illustrating genetic relationships among 36 major cereal crops (rice, wheat, maize, barley, pearl millet and sorghum) genotypes, generated by UPGMA cluster tree analysis.
Conclusion
This reveals a relatively high degree of genetic variability within the two cultivars. The highest pairwise similarity index was between Pearl Millet (GP1) and Sorghum (CSV-21(F)) (0.81) (Table 3) indicating that these are relatively less variable. The data obtained in this study confirmed the efficiency of the RAPD technique as a valuable DNA marker for determination and estimation of genetic similarity among different plant genotypes. The information about genetic similarity will be helpful to avoid any possibility of elite germplasm becoming genetically uniform. However, the investigation of morphological and geographical relationships of crops is also possible by RAPD analysis[23]. RAPD analysis showed good potentiality to determine phylogenetic relationships among six major cereal crops cultivars and the information about genetic similarity will be helpful to avoid any chance of elite germplasm becoming genetically uniform and endangering long term productivity gains during breeding programs. The estimation of the genetic distance between genotypes and identification of parents for performing appropriate crosses, and reaching maximum heterosis in hybridization programs. The use of RAPD markers[24] shows that this method is informative and can be used to determine the phylogenetic relationships between cultivars. Cultivars with the most distinct DNA profiles were likely to contain the greatest number of novel genes.
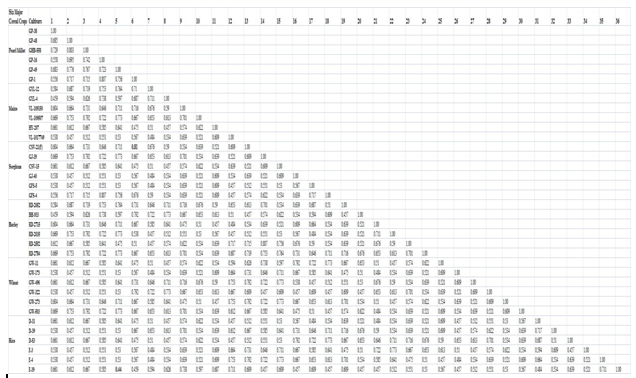
Table 3: The coefficient values of correlation among 20 RAPD primers under the genetic similarity aspect of the six major cereal crops (rice, wheat, maize, barley, pearl millet and sorghum) cultivars studied.
Figure 3: The coefficient values of correlation among 20 RAPD primers under the genetic similarity aspect of the six major cereal crops (rice, wheat, maize, barley, pearl millet and sorghum) cultivars studied.
References
- 1. Rahman, S.N., Islam, MD.S., Alam, MD.S., et al. Genetic polymorphism in rice (Oryza sativa L.) through RAPD analysis. (2007) Ind J Biotechnol 6: 224- 229.
- 2. Botstein, D., White, R.L., Skolnick, M., et al. Construction of genetic linkage map in man using restriction length polymorphisms. (1980) Am J Hum Genet 32(3): 314- 331.
- 3. Tanysley, S.D., Young, N.D., Paterson, A.H., et al. RFLP mapping in plant breeding: New tools for an old science. (1989) Nat Biotechnol 7: 257- 264.
- 4. Williams, J.G., Kubelik, A.R., Livak, K.J., et al. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. (1990) Nucleic Acids Res 18(22): 6531- 6535.
- 5. Vos, P., Hogers, R., Bleeker, M., et al. AFLP: a new technique for DNA fingerprinting. (1995) Nucleic Acids Res 23(21): 4407- 4414.
- 6. Tautz, D. Hypervariablity of simple sequences as a general source of polymorphic DNA markers. (1989) Nucleic Acids Res 17(16): 6463- 6471.
- 7. Hadrys, H., Balick, M., Schierwater, B. Application of random amplified polymorphic DNA (RAPD) in molecular ecology. (1992) Mol Ecol (1): 55- 63.
- 8. Thomas, G., Mohapatra, T., Rao, A. R., et al. Distinguishing Indian commercial wheat varieties using RAPD based DNA fingerprints. (2006) Ind J Biotechnol (5): 200- 206.
- 9. Michelmore, R.W., Paran, I., Kesseli, R.V. Identification of markers linked to disease-resistance genes by bulked segregante analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. (1991) Proc Natl Acad Sci U.S.A 88(21): 9828- 9832.
- 10. Haig, S.M., Rhymer, J.M., Heckel, D.G. Population differentiation in randomly amplified polymorphic DNA of red cockaded woodpeckers Picoides borealis. (1994) Mol Ecol 3(6): 581- 595.
- 11. Chapco, W., Ashton, N.W., Martel, R.K., et al. A feasibility study of the use of random amplified polymorphic DNA in the population genetics and systemic of grasshoppers. (1992) Genome 35(4): 569- 574.
- 12. Raghunathachari, P., Khanna, V.K., Singh, U.S., et al. RAPD analysis of genetic variability in Indian scented rice germplasm (Oryza sativa L.). (2000) Current Science (79): 994- 998.
- 13. Prakash, S.P.J., Biji, K.R., Gomez, S.M. Genetic diversity analysis of sorghum (Sorghum bicolor L. Moench) accessions using RAPD markers. (2006) Ind J Crop Sci 1(1-2): 109-112.
- 14. Govindaraj, M., Selvi, B., Arun Prabhu, D., et al. Genetic diversity analysis of pearl millet (Pennisetum glauccum [L.] R. Br.) Accessions using molecular markers. (2009) African J Biotechnol 8(22): 6046-6052.
- 15. Kachapur, R.M., Salimath, P.M., Reddy, B.V.S., Genetic Diversity within Sweet Sorghum (Sorghum bicolor (L.) Monech) Accessions as Revealed by RAPD Markers. (2009) Journal of Maharashtra Agricultural Universities 34(1): 38-42.
- 16. Kumar, R., Manish, J.K., Vishwakarma, K., et al. Assessment of genetic diversity and its usefulness for varietal identification in Indian elite varieties of wheat (T. aestivum L.) Using RAPD Markers. (2011) Asian J Biotechnol 3(5): 460-469.
- 17. Suresh, H., Sasidharan, N., Sudeshna, C., et al. Genetic diversity among maize varieties revealed by phenotypic descriptors and rapd profiles. (2013) J Agricult Sci 8(2): 91-106.
- 18. Agrama, H.A., Tunistra, M.R. Phylogenetic diversity and relationship among sorghum accession using SSR’s and RAPD’s. (2003) African J Biotechnol 2(10): 334-340.
- 19. Doyle, J.J., Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. (1987) Phytochemi Bulletin (19): 11-15.
- 20. Nei, M., Li, W.H. Mathematical model for studying genetic variation in terms of restriction endonucleases. (1979) Proc Natl Acad Sci U. S. A 76(10): 5269-5273.
- 21. Rohlf, F.J. NTSYS-pc: Numerical Taxonomy and Multivariate Analysis System, Version 2.2. New York. (1992) Exeter Publications
- 22. Powell, W., Morgante, M., Andre, C., et al. The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. (1996) Molecular Breeding (2): 225-238.
- 23. Ashraf, M., Afsari, S., Abdul Gafoor, Q. Total DNA variation into varieties using known primers of the genes induced in dehydration and salinity. (2003) Pak J Biol Sci 6(5): 437- 440
- 24. Paterson, A.H., Tanksley, S.D., Sorrels, M.E. DNA markers in plant improvement. (1991) Advan Agron (46): 39-90.