Implication of Hsp70 Gene Polymorphisms in Arrhythmogenic Right Ventricular Cardiomyopathy/Dysplasia
C. Sandeep Kumar1, M.L. Satynarayana1, Ananthapur Venkateshwari2, Calambur Narasimhan3, Pratibha Nallari1*
Affiliation
- 1Department of Genetics, University College of Science, Osmania University, Hyderabad, Telangana, India
- 2Institute of Genetics and Hospital for Genetic Diseases, Osmania University, Hyderabad, Telangana, India
- 3Care Hospital, Hyderabad, Telangana, India
Corresponding Author
Pratibha Nallari, Professor, Department of Genetics, Osmania University, College of Science, Hyderabad-500007, India. Tel: 91-040- 27682335; Fax: 91-040-27098020; E-mail: prathinallari@gmail.com
Citation
Nallari, P., et al. Implication of HSP70 gene polymorphisms in Arrhythmogenic right ventricular cardiomyopathy/dysplasia. (2015) J Heart Cardiol 1(1): 14-18.
Copy rights
© 2015 Nallari, P. This is an Open access article distributed under the terms of Creative Commons Attribution 4.0 International License
Keywords
Arrhythmogenic right ventricular cardiomyopathy; Desmosomal genes; Modifier genes; HSP70 gene polymorphisms; Inflammatory responses; Apoptosis
Abstract
Introduction: Arrhythmogenic right ventricular cardiomyopathy/dysplasia (ARVC/D) is characterized by initial fibrofatty replacement of the right ventricle. Mutations in the Desmosomal and non desmosomal apart from modifier genes are known to be involved in the pathogenesis of the ARVC/D. One such modifier gene identified is HSP70 which plays an important role in cardio-protection. The present study investigates the role of Hsp70 polymorphisms in ARVC/D disease severity.
Methods: Analysis of 100 control samples and 33 ARVC/D patients for 3 polymorphic loci was carried out by PCR - RFLP. Statistical analysis was carried out by SNPSTAT and haploview to analyze the genotypic data generated.
Results: Our study revealed a statistically significant association of GC genotype (HSP70-1 +190G/C) [OR 2.93,95%CI 1.08-8.00, p = 0.035] and TC genotype (HSP70-hom +2437T/C) [OR 2.04, 95% CI 1.01-4.55, p = 0.045] with risk to develop ARVC/D. The haplotype frequency of CGT was also found to be higher in patients compared to controls, strengthening the synergistic effect of HSP70 family in ARVC/D.
Conclusions: The present investigation revealed the importance of HSP70 polymorphisms in the pathogenesis of the ARVC/D. The results highlight a modifier role of HSP70 polymorphisms in ARVC/D etiopathogenesis, as the synergistic action of these polymorphisms may inhibit the HSP70 protein to perform its regular role, which in turn may trigger inflammation and apoptosis, hence the muscle tissue may be replaced by fibrofatty tissue.
Introduction
Arrhythmogenic right ventricular cardiomyopathy/dysplasia (ARVC/D) is a rare multifactorial disorder, characterized by fibrofatty replacement of right ventricle. The replacement of myocytes in the right ventricle by fibrous tissue leads to right ventricular failure, arrhythmias, and sudden cardiac death. The prevalence of this cardiomyopathy varies from population to population with an average estimate being 1in 5000/live births. Males are more frequently affected than females with a ratio of 3:1. It can be inherited in an autosomal dominant pattern with variable expression and reduced penetrance. In some forms, autosomal recessive pattern of inheritance is also reported. Till date different desmosomal and non-desmosomal genes have been implicated in the pathophysiology of ARVC/D (Romero et al, 2013).
However, the phenotype depends on genetic background of an individual, in addition to the causal mutation; hence modifier genes seem to influence the severity of the phenotypic expression and this is especially seen in autosomal dominant disorders like cardiomyopathy (Slavotinek and Biesecker, 2003).
One of the modifiers is the cardioprotective HSP70 gene family, a highly conserved gene family and expression is enhanced on exposure to stressful stimuli. It has wide range of protective effects like enhancing the immune responses, thermotolerance and cytoprotection from inflammatory mediators (Smith et al, 2007). HSP70 gene family consists of three variants viz HSP70-1, HSP70-2 and HSP70-hom and is mapped to the MHC class III region on chromosome 6 p23.1. Polymorphisms in HSP70 have been implicated as a risk factor in several disorders Parkinson's disease (Wu et al, 2004), high-altitude illness (Zhou et al, 2005), aging (Singh et al, 2006a, b), and uveitis (Spagnolo et al, 2007). Since ARVC/D is characterized by inflammation of right ventricle; involvement of HSP70 polymorphisms as a cytoprotective modifier, is therefore, examined in the present study.
Materials and Methods
The 2010 diagnostic criteria were used to establish the diagnosis of ARVC/D. Definite ARVC/D was characterized by the presence of ≥2 major criteria, 1 major and 2 minor criteria or 4 minor criteria (Marcus et al, 2010). ARVC/D is one of the rarest disorders reported to date.
33 unrelated probands and 100 healthy control blood samples were collected from Cardiology Unit of CARE Hospital, Hyderabad and Osmania General Hospital, Hyderabad respectively, after obtaining written informed consent. The study was conducted according to guidelines of local institutional ethical committee clearance from CARE hospitals, Hyderabad was also obtained. DNA isolation was carried out following the methodology as described by (Lahiri and Nurnberger, 1991).
Molecular analysis
Genotypes of HSP-70-1+190G/C (rs1043618), HSP-70-2+1267A/G (rs1061581), and HSP-70-hom +2437T/C (rs2227956), were determined by PCR based RFLP analysis. The PCR reaction was carried out in 0.2ml tubes with 25μl of master mix and each tube contained 100ng of genomic DNA, 50p moles each of forward and reverse primer, 200μm of dNTPs, 1X PCR buffer. The conditions applied are: Initial denaturation at 95°C for 3 min, followed by denaturation at 95°C for 30s, annealing at 55°C-58°C (depending on the exon) for 30s, extension at 72°C for 1min and final extension at 72°C for 2 min for 30 cycles. The amplicons were subjected to RFLP analysis by the respective restriction enzymes and incubated overnight and were later checked on 10% acrylamide gels. The primer sequences procured were based on Milner and Campbell, (1992).Table 1
Table 1: Primers and restriction enzymes as described by Milner and Campbell (1992)
| Polymorphism | Chromosome | Primers | Annealing temperature /Mgcl2mM | Amplifier | Enzyme | RFLP Base pairs |
|---|---|---|---|---|---|---|
| HSP-70-1+190G/C | 6 | F:CGCCATGGAGACCAACACCC R:GCGGTTCCCTGCTCTCTGTC | 55°C/1.0 | 488 bp | BsrBI | 461+27 |
| HSP-70-2+1267A/G | 6 | F:CATCGACTTCTACACGTCCA R:CAAAGTCCTTGAGTCCCAAC | 58°C/2 | 1118 bp | Pst-1 | 934+184 |
| HSP-70-hom +2437T/C | 6 | F: GTCCCTGGGGCTGGAGACGG R:GATGATAGGGTTACACATCTGCT | 57°C/1.0 | 627bp | NcoI | 354+273 |
Statistical analysis
Genotypic and allelic frequencies were calculated for each polymorphic locus. Odds ratios with 95% confidence intervals (95%CI) were estimated to test for relative risk of association between various genotypic combinations by using SNPStat software (Solé et al, 2006). The difference in the allelic and genotypic frequencies of controls and patient group were tested by chi square test for statistical inference. A hapmap analysis was carried out with respect to the allelic frequencies of other population groups available on international hapmap project (NCBI). Haplotype analysis and linkage disequilibrium analysis were carried out by haploview 4.2 software (Barrett et al, 2004) to examine the association between various polymorphisms of HSP70 gene family. P values < 0.05 was considered as statistically significant.
Results
The control and patient DNA samples were genotyped for the 3 polymorphic loci. The genotypic and allele frequencies and odds relative risk estimation for three polymorphic loci are present in Tables 2- 6.
Table 2: Allelic distribution of HSP70 gene polymorphisms.
| Polymorphic sites | Alleles | Controls frequency | ARVC/D frequency |
|---|---|---|---|
| HSP70 1 | +190 G/C | ||
| G | 0.58 | 0.52 | |
| C | 0.42 | 0.48 | |
| HSP70 1 | +1267A/G | ||
| A | 0.58 | 0.55 | |
| G | 0.42 | 0.45 | |
| HSP70 hom | +2437T/C | ||
| T | 0.60 | 0.55 | |
| C | 0.40 | 0.45 | |
Table 3: Genotypic distribution of HSP70 gene polymorphisms
| Polymorphic sites | Alleles | Controls | ARVC/D | ||
|---|---|---|---|---|---|
| HSP70-1 | +190 G/C | N | % | N | % |
| GG | 36 | 36 | 6 | 18 | |
| GC | 45 | 45 | 22 | 67 | |
| CC | 19 | 19 | 5 | 15 | |
| HSP70-2 | +1267A/G | ||||
| AA | 33 | 33 | 10 | 30 | |
| AG | 51 | 51 | 16 | 49 | |
| GG | 16 | 16 | 7 | 21 | |
| HSP70-hom | +2437T/C | ||||
| TT | 38 | 38 | 8 | 24 | |
| TC | 43 | 43 | 20 | 61 | |
| CC | 19 | 19 | 5 | 15 | |
Table 4: Genotypic frequencies and relative risk estimates of HSP70-1 patients and control
| Model | Genotype | Controls N (%) | ARVC/D N (%) | OR (95%CI) | p-value |
|---|---|---|---|---|---|
| Codominant | GG | 36 (36) | 6 (18) | 1.00 | |
| GC | 45 (45) | 22 (67) | 2.93 (1.08-8.00) | 0.035 | |
| CC | 19 (19) | 5 (15) | 1.58 (0.43-5.85) | 0.49 | |
| Dominant | GG | 36 (36) | 6 (18) | 1.00 | |
| GC+CC | 64 (64) | 27 (82) | 2.53 (0.96-6.71) | 0.048 | |
| Recessive | GG+GC | 81 (81) | 28 (85) | 1.00 | |
| CC | 19 (19) | 5 (15) | 0.76 (0.26-2.23) | 0.61 | |
| Overdominant | GG+CC | 55 (55) | 11 (33) | 1.00 | |
| GC | 45 (45) | 22 (67) | 2.44 (1.07-5.57) | 0.03 |
Table 5: Genotypic frequencies and relative risk estimates of HSP70-2 patients and control
| Model | Genotype | Control N (%) | ARVC/D N (%) | OR (95% CI) | P-value |
|---|---|---|---|---|---|
| Codominant | AA | 33 (33) | 10 (30) | 1.00 | |
| AG | 51 (51) | 16 (49) | 1.04 (0.42-2.55) | 0.94 | |
| GG | 16 (16) | 7 (21) | 1.44 (0.46-4.49) | 0.52 | |
| Dominant | AA | 33 (33) | 10 (30) | 1.00 | |
| AG+GG | 67 (67) | 23 (70) | 1.13 (0.48-2.65) | 0.77 | |
| Recessive | AA+AG | 84 (84) | 26 (79) | 1.00 | |
| GG | 16 (16) | 7 (21) | 1.41 (0.52-3.81) | 0.5 |
Table 6: Genotypic frequencies and relative risk estimates of HSP70-hom patients and control
| Model | Genotype | Controls N (%) | ARVC/D N (%) | OR (95% CI) | P-value |
|---|---|---|---|---|---|
| Codominant | TT | 38 (38) | 8 (24) | 1.00 | |
| TC | 43 (43) | 20 (61) | 2.21 (0.87-5.59) | 0.094 | |
| CC | 19 (19) | 5 (15) | 1.25 (0.36-4.34) | 0.72 | |
| Dominant | TT | 38 (38) | 8 (24) | 1.00 | |
| TC+CC | 62 (62) | 25 (76) | 1.92 (0.78-4.68) | 0.15 | |
| Recessive | TT+TC | 81 (81) | 28 (85) | 1.00 | |
| CC | 19 (19) | 5 (15) | 0.76 (0.26-2.23) | 0.61 | |
| Overdominant | TT+CC | 57 (57) | 13 (39) | 1.00 | |
| TC | 43 (43) | 20 (61) | 2.04 (1.01-4.55) | 0.046 |
+190G/C of HSP70-1
Statistical analysis of HSP70-1 revealed that 'GC' genotype was significantly higher in cases (67%) when compared to control group (45%). The relative odds ratio points towards a predisposing effect of 'GC' genotype with about 3 fold increased risk to ARVC/D [OR 2.93, 95% CI 1.08-8.00, p=0.035]. The association of 'GC' genotype with the disease progression holds well when tested against both the homozygote's (TT+CC) in overdominant model.
+1267A/G of HSP70-2
There is no significant difference observed in the frequency distribution of genotypes in both control and patient samples with respect to the HSP70-2 +1267A/G locus.
+2437T/C of HSP70-hom
In case of HSP70-hom +2437T/C polymorphism, 'TC' genotype was significantly higher in patients (60%) compared to control group (43%), with 2 fold increased risk to ARVC/D [OR 2.04, 95% CI 1.01-4.55, p=0.045], strengthening the selective disadvantage of heterozygotes in cases of ARVC/D.
Hapmap analysis
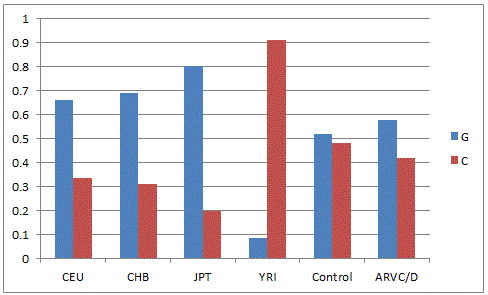
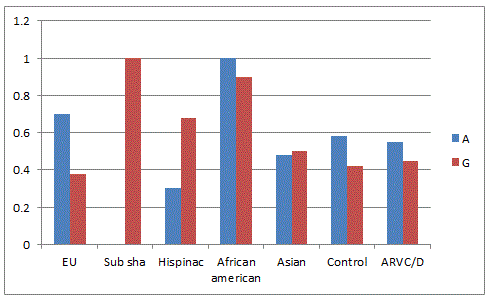
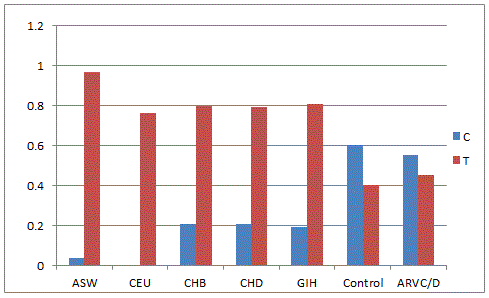
Hapmap analysis was carried out by comparing the allele frequencies of global population present in the dbSNP database (NCBI) with the present hapmap of HSP70-1(Figure-1) the 'G' allele was found to be dominant in Utha residents, Han Chinese, Japanese and Yourban in Nigeria while in the Japanese population, the 'C' allele was found to be pre-dominant. In the Indian control population, both alleles are nearly equal but in patient cohort, it was nearly one fold higher. The'A' allele of HSP70-2 (Figure-2) was predominant in European, African and Indian population whereas 'G' allele was higher in Hispanic and Subsharan American population and cohort of Indian origin. The allelic comparison of HSP70-hom (Figure-3) in different population revealed the higher frequency of 'T' allele in African ancestry, Utah residents, Hans Chinese and Gujarati Indians in Houston but Indian population has higher 'C' allele contributing to ethnicity and genetic diversity.
Linkage disequilibrium and haplotype analysis
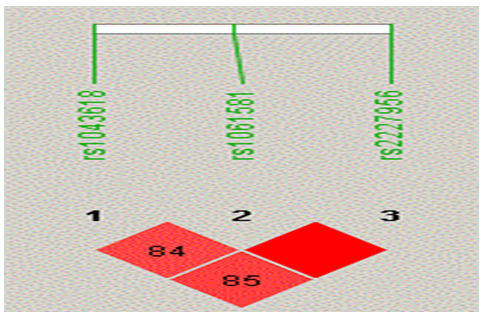
Linkage disequilibrium (LD) and haplotype analysis was carried out for the 3 genotypes of HSP70 using Haploview 4.2 software, to examine for any possible association with the disease phenotype. Pairwise LD comparison of the three polymorphisms in patient and control groups, depicting the measures is represented in Figure 4 and 5 respectively. Significant D' values were observed for HSP70-1 +190 G/C (rs1043618) and HSP70-2 +1267A/G [(rs1061581) (D'=0.84)], HSP70-1 +190 G/C and HSP70-hom (0.85) and HSP70-2 +1267A and HSP70-hom (rs2227956) (1.00) indicating a tight linkage disequilibrium between the SNPs (Figure-4). Pairwise comparison of these 3 polymorphic loci in control samples revealed a perfect LD (Figure-5).
Haplotype based Linkage disequilibrium mapping has become a powerful and cost effective tool for performing genetic association studies. Our results from haplotype analysis revealed that the haplotypic frequencies of CAT and CGT were significantly higher in patients when compared other haplotypes. CGT haplotype was found to confer higher risk to ARVC/D with significant odds ratio.
Linkage disequilibrium and haplotype analysis
Linkage disequilibrium (LD) and haplotype analysis was carried out for the 3 genotypes of HSP70 using Haploview 4.2 software, to examine for any possible association with the disease phenotype. Pairwise LD comparison of the three polymorphisms in patient and control groups, depicting the measures is represented in Figure 4 and 5 respectively. Significant D' values were observed for HSP70-1 +190 G/C (rs1043618) and HSP70-2 +1267A/G [(rs1061581) (D'=0.84)], HSP70-1 +190 G/C and HSP70-hom (0.85) and HSP70-2 +1267A and HSP70-hom (rs2227956) (1.00) indicating a tight linkage disequilibrium between the SNPs (Figure-4). Pairwise comparison of these 3 polymorphic loci in control samples revealed a perfect LD (Figure-5) & Table 7.
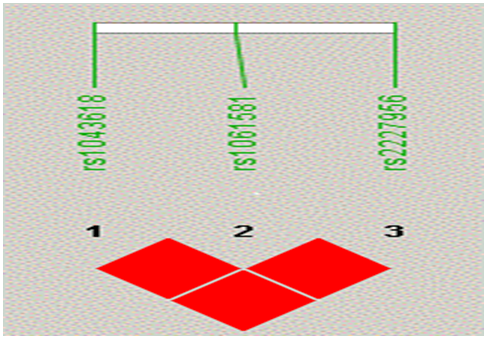
Figure 5: Pairwise Control samples LD showing perfect LD between three HSP70 (1, 2 and hom) polymorphisms
Table 7: Haplotype frequency distribution and association with patient group compared to Controls
| HSP70-1 | HSP70-2 | HSP70-hom | Control | ARVC/D | OR (95% CI) | p-value |
|---|---|---|---|---|---|---|
| G | G | T | 0.415 | 0.4207 | 1.00 | --- |
| C | A | C | 0.405 | 0.4204 | 1.12 (0.60 - 2.09) | 0.73 |
| G | A | T | 0.17 | 0.0603 | 0.33 (0.10 - 1.09) | 0.072 |
| C | A | T | 0.01 | 0.0306 | 2.43 (0.26 - 23.05) | 0.44 |
| C | G | T | 0 | 0.0339 | 44379451.15 (44379442.75 - 44379459.54) | < 0.0001 |
Haplotype based Linkage disequilibrium mapping has become a powerful and cost effective tool for performing genetic association studies. Our results from haplotype analysis revealed that the haplotypic frequencies of CAT and CGT were significantly higher in patients when compared other haplotypes. CGT haplotype was found to confer higher risk to ARVC/D with significant odds ratio.
Discussion
To date, research on ARVC/D revealed the involvement of desmosomal and non desmosomal gene/s variation (SNPs) and environmental factors. The phenotypic expression of clinical condition depends on both gene variants and genetic modifiers, which may influence the severity and progression of the condition.
One of the well-established gene modifiers is the HSP70 gene family, which is a highly conserved gene family. The expression is enhanced during stress stimuli, indicating their role in cell repair. Their important role as the regulators of the pathophysiology and cardioprotection in heart-related diseases, cancer, diabetes and aging is well established. As a part of cell protective mechanism sequence variations of HSP70 can alter protein function and structure leading to variations in tolerance to stress which may enhance susceptibility to the diseases. HSP70 was also found in intracellular compartments of cells and these proteins are involved in the following important functions 1)folding and assembly of nascent polypeptides 2) refolding of misfolded or aggregated proteins 3) promoting the ubiquitination and degradation of misfolded proteins 4) preventing protein aggregation 5) Apoptosis and 6) Immune responses (Turturici et al, 2010).
To our knowledge, this is the first study investigating the role of HSP70 gene in the pathogenesis of ARVC/D. It is hypothesized that alteration in the protein structure of HSP70 due to genetic variations may influence the subsequent release of cell components by necrosis in circulation, thereby increasing the risk to inflammation as a pro-inflammatory modulator in ARVC/D.
The present study revealed the ('GC' genotype of +190 G/C HSP70-1 and TC +2437 T/C HSP70-hom) to be associated with ARVC/D. These polymorphisms may alter the repairing capability of HSP70 by accumulation of misfolded proteins in cardiac cells and tissue, which is corroborated by the significant role of HSP70-1 and HSP70-hom polymorphism of the HSP70 gene family.
HSP70-1 Polymorphism
The +190 G/C HSP70-1 polymorphism is a silent substitution in the 5' flanking and noncoding region of both the constitutively expressed and heat-inducible form.
Endogenous role of HSP70 and ARVC/D pathogenesis
Induced HSP70 proteins are key components of endogenous pathway that limit the extent of myocardial damage caused by stress. They also play an important role in the maintenance of integrity of the cytosolic proteins; enhance the survival of cardiomyocytes by inhibiting the pro-apoptotic pathways (Fas mediated death cascade and caspase dependent pathway) and conservation of anti-oxidants. HSP70 residing in the native compartment has the ability to hamper ongoing pro-inflammatory signaling cascade and promote down regulation of inflammatory responses by targeting the NF-κB pathway (De Jong PR et al, 2009). Since ARVC/D is associated with apoptosis wherein cardiomyocytes are replaced by fibrofatty replacement in RV of the heart, such an association can be justified.
HSP70 mediated negative regulation involve inhibition of IκB kinase (IKK) activity by interacting with IKK gamma subunit. Alternatively the interaction and switching of tumor necrosis factor receptor associated factor 6 (TRAF6), an important molecule in TLR signaling, may subsequently activate NF-κB leading to initiation of inflammation (Guzhova et al, 1997). It is hypothesized that loss of interaction of HSP70 with NF-κB and its associated proteins leads to initiation of an inflammatory response and subsequent ventricular remodeling involving fibrofatty replacement followed by sudden cardiac death. Hence, loss of normal protein interaction network of HSP70 may result in ARVC/D and justifies the role of HSP70 gene family as a modifier of disease phenotype.
Thus, altered HSP70 with 'GC' (risk) genotype in myocardium may be unable to carry out its normal function and lead to the loss of endogenous control of above essential pathways, required for the cell survival, cell death and fibrofatty replacement of myocardial ventricles.
HSP70-hom +2437T/C
HSP70-hom +2437T/C is a missense mutation Met-439Thr, which may affect HSP70-hom protein folding and subsequent other protein foldings in the cell. According to Pociot's theory, a methionine substitution by threonine at 439 residue may affect which HSP70- hom's functional ability as molecular chaperone by decreasing the hydrophobic interactions to its target unstructured protein (Pociot et al, 1993). Dysfunctional proteins may disrupt the cellular membrane formation process. Hence, altered proteins lead to the defective cell membrane and release of cell contents including altered HSP70-hom which acts as co-inducer to innate immune system as it has a protective role by inhibiting apoptosis and anti-inflammatory responses.
The exact mechanism by which HSP-hom inhibits apoptosis is still unknown. But HSP-hom can interact and interfere with pro-apoptotic protein responsible for apoptosis activation possible mechanisms include 1) HSP70 interacts with Apaf-1 and inhibit the activation of caspases 2) It directly interacts with Bax and suppresses its activation 3) HSP70 inhibits the apoptotic pathway downstream of caspase-3 activation and 4) chromatin condensation induced by apoptosis inducing factor (Jaattela et al, 1998 ).
HSP70-hom can have cytoprotective effect by suppressing the activation of NF-κB, NF- κB triggers the production of various pro-inflammatory cytokines such as IL-1β, IL-6 and TNF-α. Interestingly, in the present study, the heterozygotes (TC) of HSP70-hom were found to be at high risk towards ARVC/D. Hence, the altered HSP70 may fail to suppress the production of pro-inflammatory cytokines which may lead to inflammation and fibrofatty replacement.
LD and Haplotype association analysis
The CGT haplotype was observed only in ARVC/D group, which is the risk conferring haplotype towards disease progression. The significant association of the CGT haplotype with ARVC/D indicates a functional role and synergistic effect of HSP70 family in ARVC/D which could be explained based on altered cardioprotection, chaperoning and/or apoptosis and inflammatory responses.
Conclusion
This is the first investigation, associating the relationship between the HSP70 polymorphisms and ARVC/D in Indian population. Statistically significant results are obtained with respect to 'GC' and 'TC' genotypes of HSP70-1 and HSP70-hom respectively, pointing the predisposing roles towards ARVC/D. Haplotype analysis revealed a possible functional and synergistic effect of HSP70 polymorphisms in the ARVC/D susceptibility. These results point towards a modifier role of HSP70 polymorphisms in ARVC/D etiopathogenesis.
Acknowledgement
My team thanks to patients and controls who donated their blood for this study. Mr. C. Sandeep Kumar is a recipient of SRF from University Grants Commission (UGC).
References
- 1. Romero, J., Eliany, M.L., Carlos M., et al. Arrhythmogenic Right Ventricular Cardiomyopathy (ARVC/D): A Systematic Literature Review. (2013) Clin Med Insights Cardiol 7: 97–114.
- 2. Slavotinek, A., Biesecker, L.G. Genetic modifiers in human development and malformation syndromes, including chaperone proteins. (2003) Human Molecular Genetics 12(Review issue 1): R45–R50.
- 3. Smith, R.S., Meyers, D.A., Peters SP., et al. Sequence analysis of HSPA1A and HSPA1B in a multi-ethnic study population. (2007) DNA Seq 18(1): 47-53.
- 4. Wu, Y.R., Wang, C.K., Chen, C.M., et al. Analysis of heat-shock protein 70 gene polymorphisms and the risk of Parkinson's disease. (2004) Hum Genet 114(3): 236–241.
- 5. Zhou, F., Wang, F., Li, F., et al. Association of Hsp70-2 and Hsp-homgene polymorphisms with risk of acute high-altitude illness in a Chinese population. (2005) Cell Stress Chaperones 10(4): 349–356.
- 6. Singh, R., Klvraa, S., Bross, P., et al. Reduced heat shock response in human mononuclear cells during aging and its association with polymorphisms in HSP70 genes. (2006)(a) Cell Stress Chaperones 11(3): 208–215.
- 7. Singh, R., Klvraa, S., Bross, P., et al. Heat-shock protein 70 genes and human longevity: a view from Denmark. (2006)(b) Ann NY Acad Sci 1067: 301–308.
- 8. Spagnolo, P., Sato, H., Marshall, S.E., et al. Association between heat shock protein 70/hom genetic polymorphisms and uveitis in patients with sarcoidosis. (2007) Invest Ophthalmol Vis Sci 48(7): 3019–3025
- 9. Marcus, F.I, McKenna, W.J., Sherrill, D., et al. Diagnosis of Arrhythmogenic right ventricular cardiomyopathy/dysplasia: proposed modification of the task force criteria. (2010) Circulation 121(13): 1533-1541
- 10. Lahiri, D., Nurnberger, J. A rapid non enzymatic method for preparation of HMW DNA from blood for RFLP studies. (1991) Nucleic Acids Res 19(19): 5444
- 11. Milner, C.M., Campbell, R.D. Polymorphic analysis of the three MHC-linked HSP70 genes. (1992) Immunogenetics 36(6): 357-362
- 12. Solé, X, Guinó, E., Valls, J. SNPStats: a web tool for the analysis of association studies. (2006) Bioinformatics 22(15): 1928-1929
- 13. Barrett, J.C., Fry, B., Maller, M., et al. Haploview: analysis and visualization of LD and haplotype maps. (2004) Bioinformatics 21(2): 263-265
- 14. Turturici, G., Sconzo, G., Geraci, F. HSP70 and Its Molecular Role in Nervous System Diseases. (2011) Biochemistry Research International 1-18
- 15. De Jong, P.R., Schadenberg, A.W., Prakken, B.J., et al. HSP70 and cardiac surgery: molecular chaperone and inflammatory regulator with compartmentalized effects. (2009) Cell Stress and Chaperones 14(2): 117–131
- 16. Guzhova, IV., Darieva, Z.A., Melo, A.R., et al. Major stress protein Hsp70 interacts with NF-kB regulatory complex in human T-lymphoma cells. (1997) Cell Stress Chaperones 2(2): 132-139
- 17. Pociot, F., Ronningen, K.S., Nerup, J. Polymorphic analysis of the human MHC-linked heat shock 70 (HSP70-2) and HSP70-hom genes in insulin-dependent diabetes mellitus (IDDM). (1993) Scand J Immunol 38(5): 491-495
- 18. Jäättelä, M., Wissing, D., Egeblad, M., et al. Hsp70 exerts its anti-apoptotic function downstream of caspase-3-like proteases. (1998) EMBO J 17(21): 6124–6134.